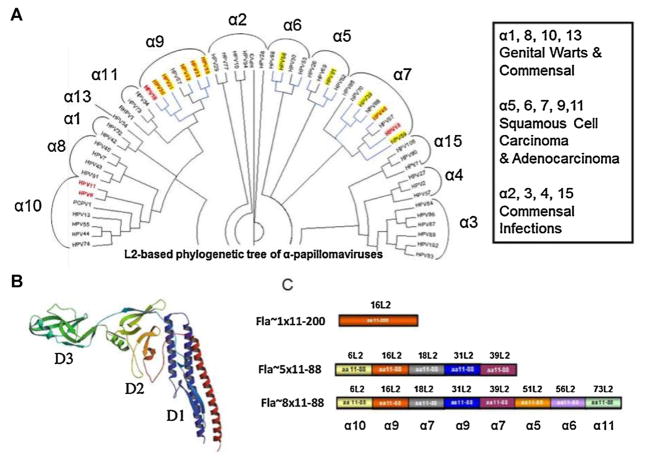
Fig. 1.
Phylogenetic tree of alphapapillomavirus and design of Fla-L2 vaccines. (A) Phylogenetic tree of alpha papillomavirus based on L2 amino acid sequences was built using the neighbor-joining Jukes-Cantor method [1]. The final tree is a consensus made from bootstrap resampling of 100 replicates. The circular representation was generated using Dendroscope [2]. Bold red font indicates HPV types represented in nona (9)-valent Merck vaccine (NCT00543543); blue lines indicate HPV types associated with cancers; pink background indicate HPV types against which licensed vaccines are ~100% protective; yellow background indicates HPV types reported to be weakly cross-protected by licensed vaccines. Alpha clade numbering is designated over the brackets. (A) (inset) – General characteristics of alpha clades (cited from Schiffman et al. [3]). (B) 3D models of Flagellin modified from [4]. Domain D1, D2, D3 domains are depicted by color. (C) A diagram showing the composition of L2 inserts for individual Fla-L2 vaccine candidates. Individual subunits were derived from different HPV species. Specific α-clades for each type represented in vaccine is depicted below Fla~8 × 11–88 L2 diagram. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)