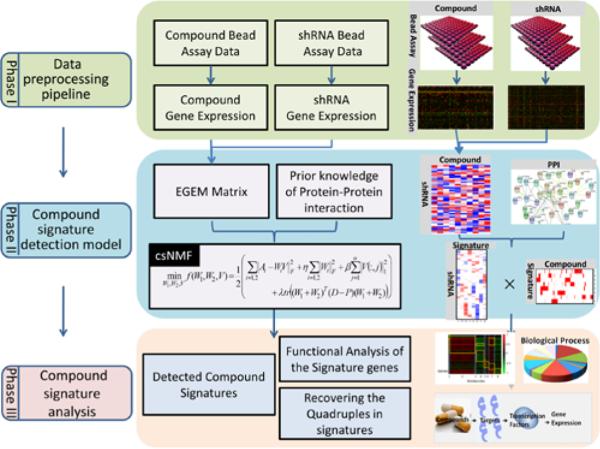
Fig. 1.
Overview of the compound signature discovery framework. This method requires raw L1000 data after various compounds and gene knockdown treatments. The raw data after the two types of treatments are preprocessed to yield gene expression data in Phase I. In Phase II, the EGEM matrix is constructed based on these gene expression data to measure relationships among compounds and knock-down genes. This matrix is then decomposed to a weight matrix and a coefficient matrix by the csNMF method. Protein-protein interaction data are added in consideration of biological connections. Signatures are identified based on strongly associated genes (i.e., those with larger values in the coefficient matrix).