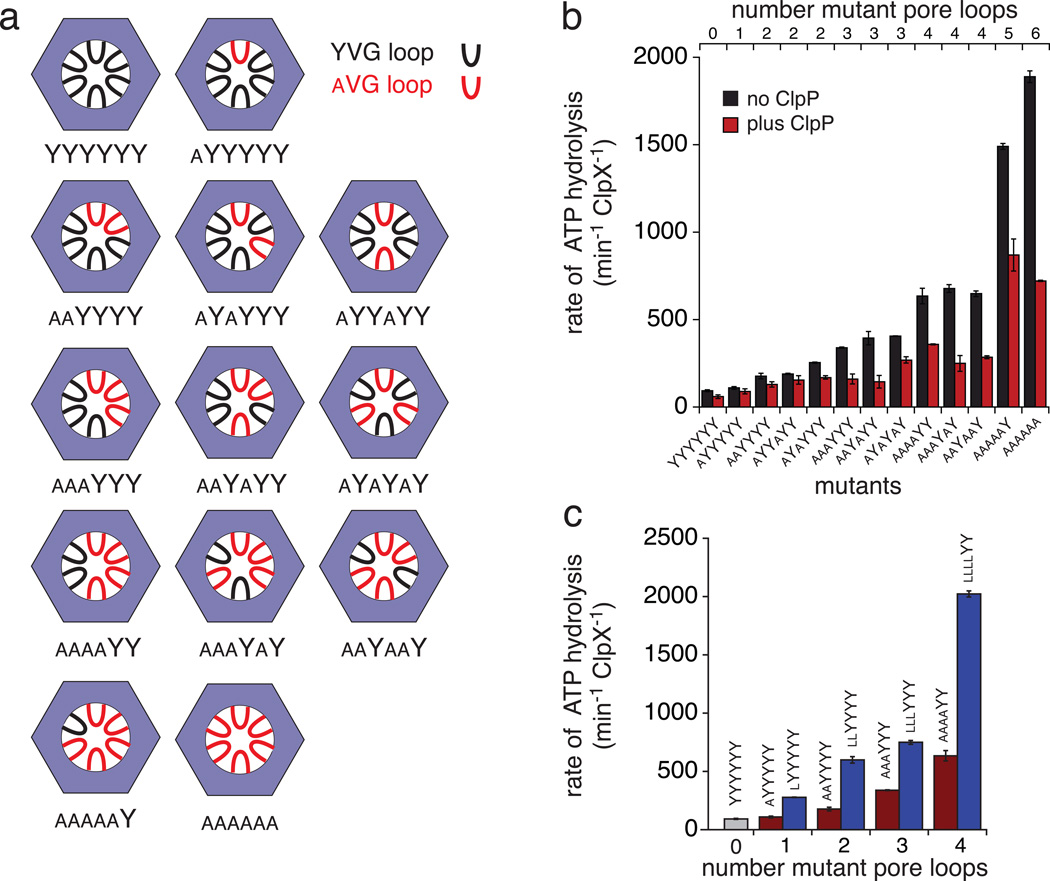
Figure 2.
Pore-loop mutants. (a) Arrangement of wild-type (YVG; black) and mutant (aVG; red) pore loops in the six subunits of covalently linked pseudo hexamers of ClpX. (b) Rates of ATP hydrolysis were measured for each pore-loop ClpX variant (100 nM) alone (dark blue) or in the presence of ClpP14 (300 nM; light red). (c) Rates of ATP hydrolysis for ClpX variants (100 nM) with one to four adjacent Y153A (dark red) or Y153L (blue) pore-loop mutations in the absence of ClpP. Error bars in panels b and c are averages (N ≥ 3) ± 1 standard deviation (SD).