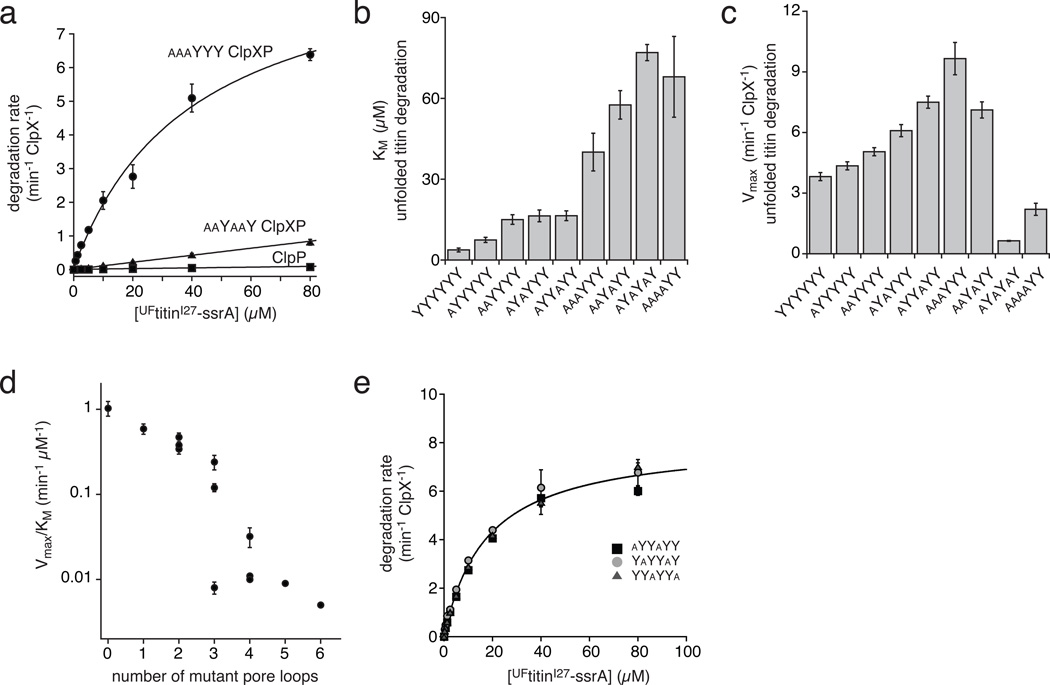
Figure 3.
Degradation of unfolded UFtitinI27-ssrA). (a) The substrate dependence of degradation rates by aaaYYY ClpXP, aaYaaY ClpXP, and ClpP alone (200 nM ClpX variant; 300 nM ClpP) were fit to the Michaelis-Menten equation for aaaYYY ClpXP and to a linear function for aaYaaY ClpXP and ClpP. Values are averages (N=3) ± 1 SD. (b) KM values for degradation of UFtitinI27-ssrA were determined from the Michaelis-Menten plots in panel a and Supplementary Fig. 1. The error of non-linear least-squares fitting is shown. (c) Vmax values and fitting errors for degradation of UFtitinI27-ssrA were determined from the Michaelis-Menten plots in panel a and Supplementary Fig. 1. (d) The second-order rate constant for UFtitinI27-ssrA ClpXP degradation (Vmax/KM) is plotted on a log scale as a function of the number of Y153A pore loop mutations in the ClpX hexamer. For variants with defined values of Vmax and KM, the error in Vmax/KM was propagated from the errors in individual parameters; for variants without defined values of Vmax and KM, the error is from linear fitting (Supplementary Fig. 1). (e) Degradation of UFtitinI27-ssrA by aYYaYY, YaYYaY, and YYaYYa ClpXP (25 nM ClpX variant; 37.5 nM ClpP), which contain the same relative configuration of wild-type and mutant pore loops in different registers with respect to the N and C termini of the single-chain hexamer. Values are averages (N=3) ± 1 SD. The line is a fit of the combined data to the Michaelis-Menten equation (Vmax = 8.1 ± 0.21 min−1 ClpX−1; KM = 17 ± 1.2 µM; R2 = 0.99).