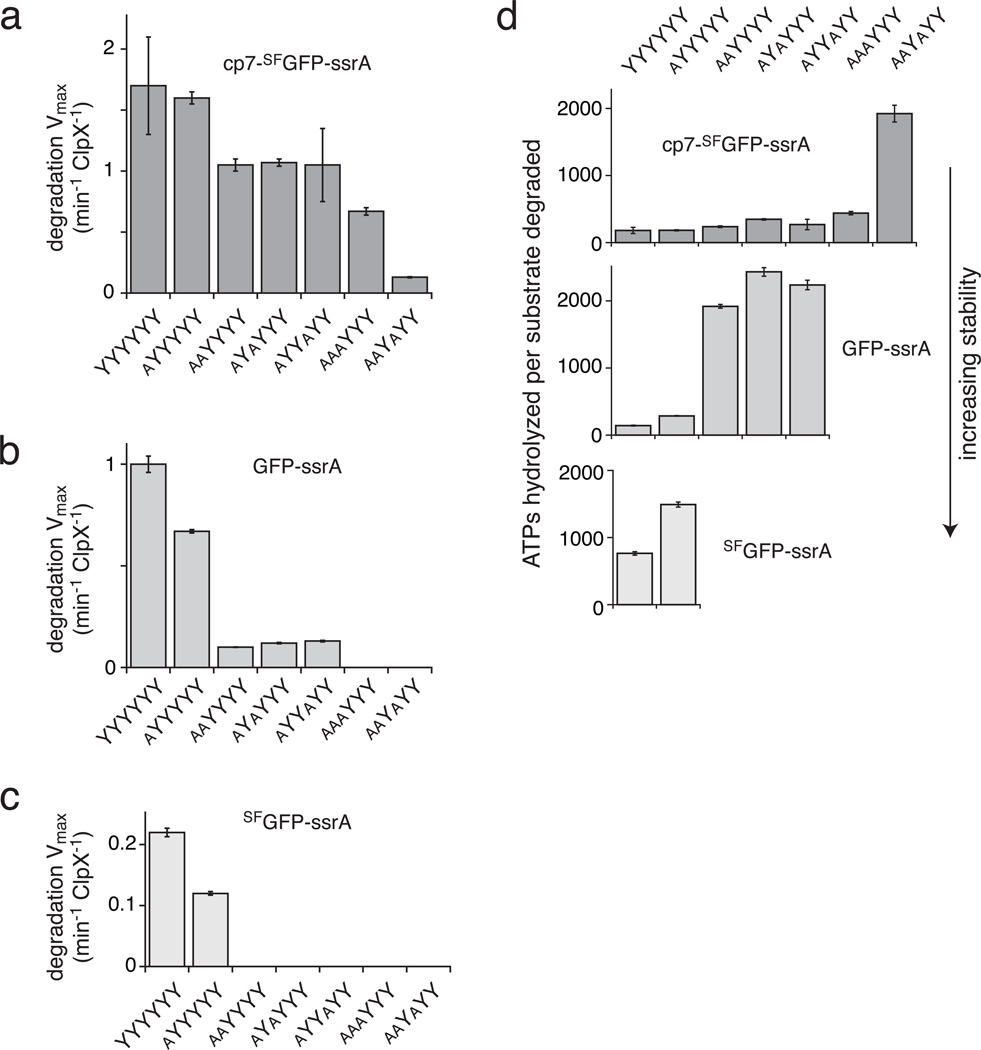
Figure 4.
Unfolding and degradation of native GFP substrates by pore-loop variants of ClpXP. (a–c) Maximal rates of degradation of cp7-SFGFP-ssrA (a), GFP-ssrA (b), and SFGFP-ssrA (c) by the YYYYYY, aYYYYY, aaYYYY, aYYaYY, aYaYYY, aaaYYY, and aaYaYY pore-loop ClpXP variants. For the enzymes that degraded these substrates, Michaelis-Menten plots are shown in Supplementary Fig. 4 and kinetic parameters are listed in Supplementary Table 2. (d) The ATP cost of degrading one molecule of native GFP substrates of increasing stability increases as the number of wild-type pore loops decreases. In all panels, values are averages (N=3) ± 1 SD.