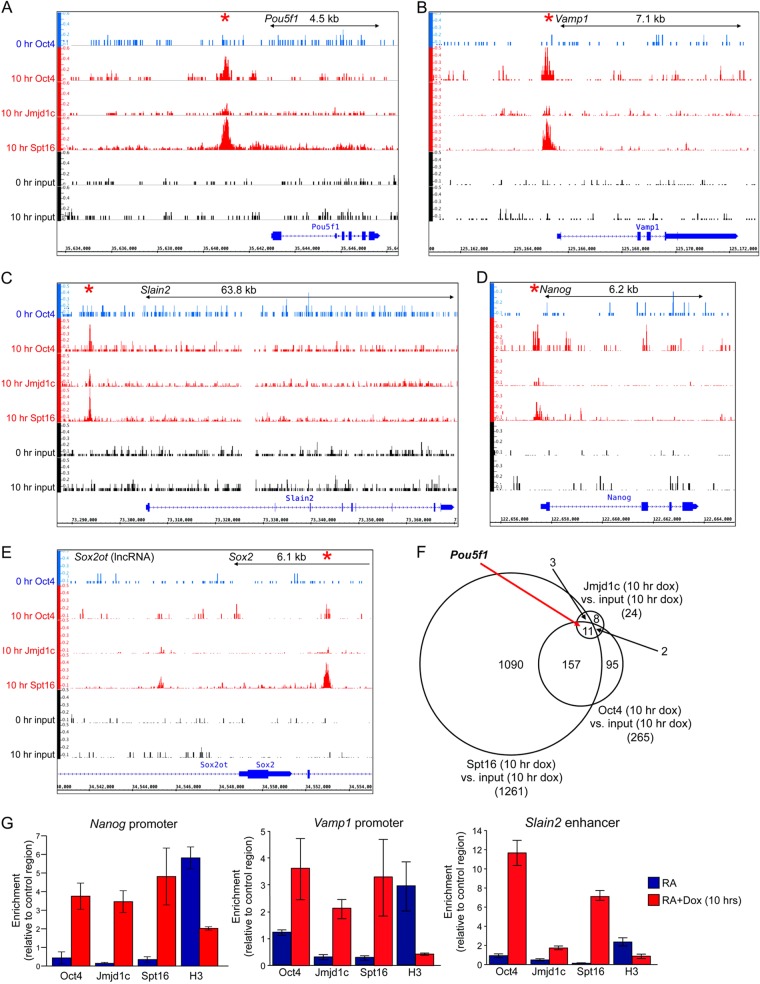
FIG 4.
Oct4, Jmjd1c, and FACT coassociate with multiple genomic targets. (A) Relative alignment depth coverage tracks of ChIP-Seq data corresponding to the murine Pou5f1 locus and surrounding region. Oct4 enrichment is shown in blue in 11-day RA-differentiated ESCs in the absence of doxycycline (0 h Oct4) or in red after a 10-h doxycycline treatment (10 h Oct4). Jmjd1c and Spt16 enrichment is shown below in red. Input controls for each time point are shown below in black. Relative depth tracks are scaled the same and represented as per-base FPKMs (fragments per kilobase of exon per million fragments). (B to E) Similar data for Vamp1 (B), Slain2 (C), Nanog (D), and Sox2 (E). A region of upstream noncoding RNA expression (Sox2ot) is also shown. (F) Venn diagram depicting target identification for Oct4, Spt16, and Jmjd1c ChIPseq, as well as overlap. The peak calling was based on a 2-fold cutoff with a Q-value FDR of 0.05. (G) ChIP enrichment for Oct4, Jmjd1c, Spt16, and histone H3 was determined by qPCR similarly to Fig. 3; however, a ChIP primer pair spanning the Oct4 site in the murine Nanog, Vamp1, and Slain2 promoters was used in place of Pou5f1.