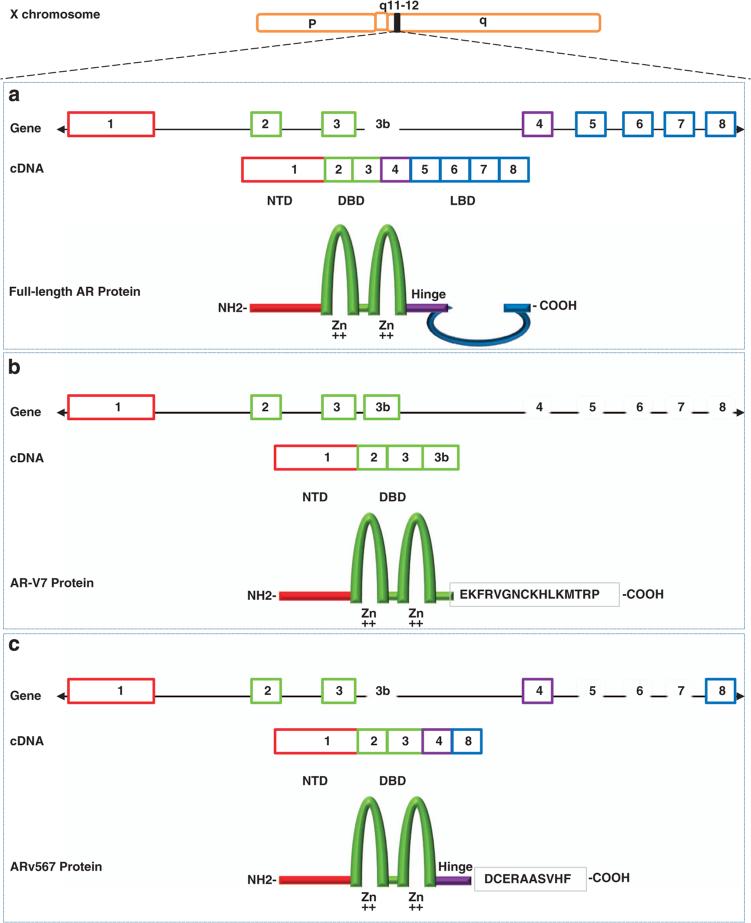
Figure 1.
Schematic structure of human AR and AR splice variant 7 (AR-V7) and 567(AR567es). (a) The human AR gene consists of eight exons with exon 1 encoding the N-terminal domain (NTD) and the entire 5′ untranslated region; exons 2 and 3 encoding the DNA-binding domain (DBD); and exons 4–8 encoding the ‘hinge’ region (HR) and ligand binding domain (LBD). The NTD contains the activation function 1 (AF-1) that includes two overlapping transcription activation units (TAU): TAU-1 (amino acids 1–370), which supports AR transcriptional activity upon stimulation by full agonist, and TAU-5 (amino acids 360–528), which confers constitutive activity to the AR in the absence of the LBD. The central region of the receptor contains the DBD and the HR and harbors the nuclear localization signal (NLS). The DBD is comprised of two cysteine-rich zinc finger motifs consisting of three alpha-helixes and a carboxy terminal extension (CTE) extending into the flexible hinge region. The first zinc finger defines DNA binding specificity, whereas the second zinc finger facilitates receptor dimerization and stabilization of the DNA-receptor complex. The carboxy-terminal end contains the LBD and the AF-2 function; (b) AR-V7 (also named AR3) encodes a protein with exons 1–3 and a terminal cryptic exon (CE3); (c) AR567es encodes a protein comprised of exons 1–4, and because of a frame-shift due to loss of exons 5–7, exon 8 has a stop codon generated after the first 10 amino acids resulting in a shortened exon 8.