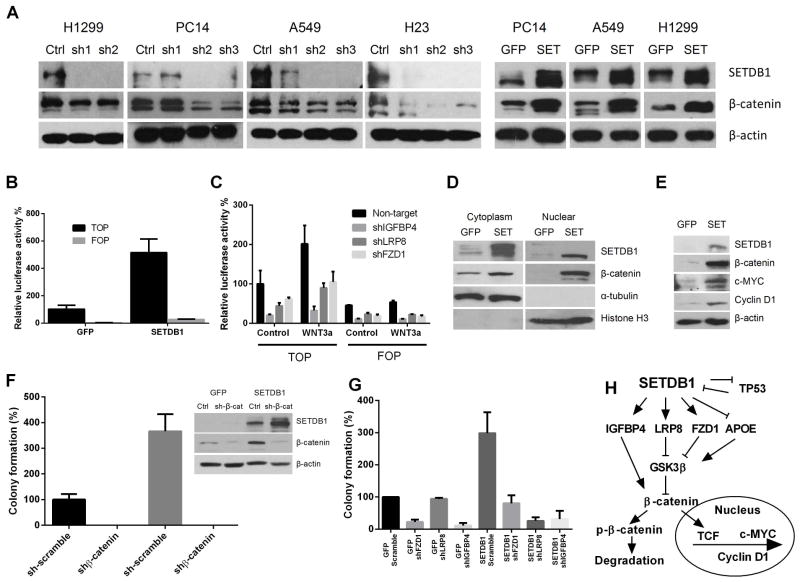
Fig. 6. Protein levels of β-catenin in the NSCLC cells with either stably overexpressed or silenced SETDB1.
A. Western analysis of β-catenin accumulation in NSCLC cell lines with either silenced or overexpressed SETDB1. Ctrl, sh-Scramble; sh1,2,3, three shRNA targeting to SETDB1. B. Relative TOP/FOP activities when overexpressing SETDB1 in H1299 cells. The stably forced expression of either SETDB1 or GFP containing cells were transfected with either pGL-TOP or pGL-FOP luciferase-reporter constructs. Luciferase activities were measured 72 hours after the transfection and normalized to the corresponding co-transfected Renilla luciferase activity. Data are shown as the ratio between TOP/FOP and Renilla. Error bars represent SD of three independent experiments. C. Silencing of IGFBP4, LRP8 or FZD1 reduced the WNT activity in H1299 NSCLC cells. Cells stably infected with either IGFBP4, LRP8 or FZD1 were transfected with either TOP or FOP, as well as Renilla control vector, and the levels of WNT pathway activity were determined by TOP/FOP assay. D. Cytoplasmic and nuclear fraction of H1299 cells stably overexpressing either GFP or SETDB1. Cells were grown on 10 cm dishes to 50% ~ 70% confluence, and the cytoplasmic and nuclear proteins were prepared as described in Materials and Methods. Cytoplasmic α-tubulin and nuclear histone H3 was used as controls of protein fractionation. E. Western analysis of alternations of downstream genes (c-MYC, Cyclin D1) of the WNT pathway in SETDB1 stably overexpressing H1299 NSCLC cells. F. Effect of stable silencing of β-catenin on clonogenic growth of H1299 cells overexpressing either GFP (control) or SETDB1. Values are expressed as % of GFP control cells, mean ± SD (three independent experiments). The silencing efficiency of β-catenin was determined by Western blot (right side). G. Clonogenic growth of either GFP or SETDB1 forced expressing H1299 cells after stable silencing of FZD1, LRP8 or IGFBP4. Values are expressed as % of GFP control cells. Experiments were performed in triplicate, and results are presented as mean ± SD. H. Schematic illustration of the proposed signaling interaction of SETDB1 and WNT pathway.