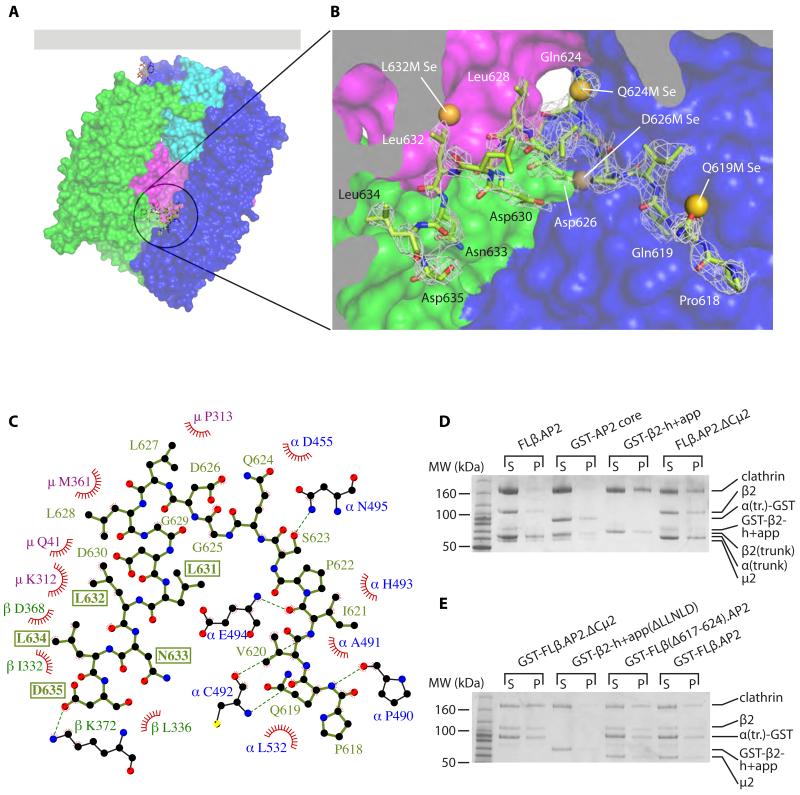
Fig. 2. The AP2 β2 subunit LLNLD clathrin binding motif is buried in the centre of the core.
(A, B). Overall (A) and closeup (B) views of the structure of βhingeHis6.AP2. The residues of the hinge resolved in the structure are shown in green as a stick representation. The AP2 core is depicted in a surface representation, coloured as in Fig. 1. The residues of the buried hinge are indicated in (B), with electron density shown as mesh (2mFo-DFc map, contoured at 0.34 e Å−3). Also shown are the positions of the selenium sites found in the bowl for each of the methionine mutants indicated, showing good agreement with the positions of the corresponding wild-type residues that were mutated. Individual LLG maps are shown in Fig S3.
(C). Ligplot+ (25) diagram showing interactions between buried hinge residues (in pale green) with residues of α (blue), μ2 (magenta) and β2 (dark green). Red fans indicate hydrophobic interactions; dashed green lines indicate hydrogen bonds. The residues of the clathrin-binding motif are boxed. See also Fig. S4.
(D). Clathrin cage assembly assays. (D) is identical to Fig. 1D (2.5 μM clathrin, 1.5 μM adaptors) with the addition of the FLβ.AP2.ΔCμ2 lane.
(E). Assays performed as in (D) but at 28°C and with 2 μM clathrin and 4 μM adaptors as shown.