Figure 1.
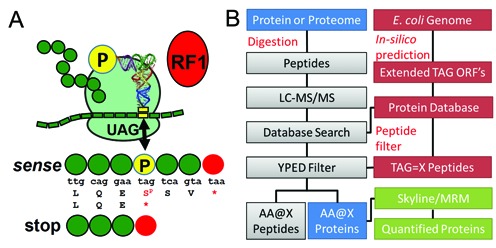
A proteomics workflow to identify natural AAs and Sep at native UAG codons in Escherichia coli. (A) Incorporation of Sep (P) at the stop codon (UAG) competes with RF1. Sep incorporation extends protein synthesis to the next in-frame stop codon (UAA or UGA) thus extending the natural ORF. (B) Proteomics workflow for the detection and quantitation of UAG stop codon readthrough. Custom open reading frame (ORF) databases are generated from the E. coli genome. ORFs are extended in silico past the natural UAG stop codon and these extended ORFs are appended to the standard E. coli proteome. These databases interface with proteomics database search software to identify extended protein ORFs from shotgun proteomics datasets. TAG sites in the extended ORFs are translated as X, or the 20 natural AAs, to populate the databases and enable peptide discovery and identification with custom filters. Protein or peptide level information can be culled from the extended proteome data and transferred into Skyline to develop MRM methods for quantitative proteomics.
