Figure 5.
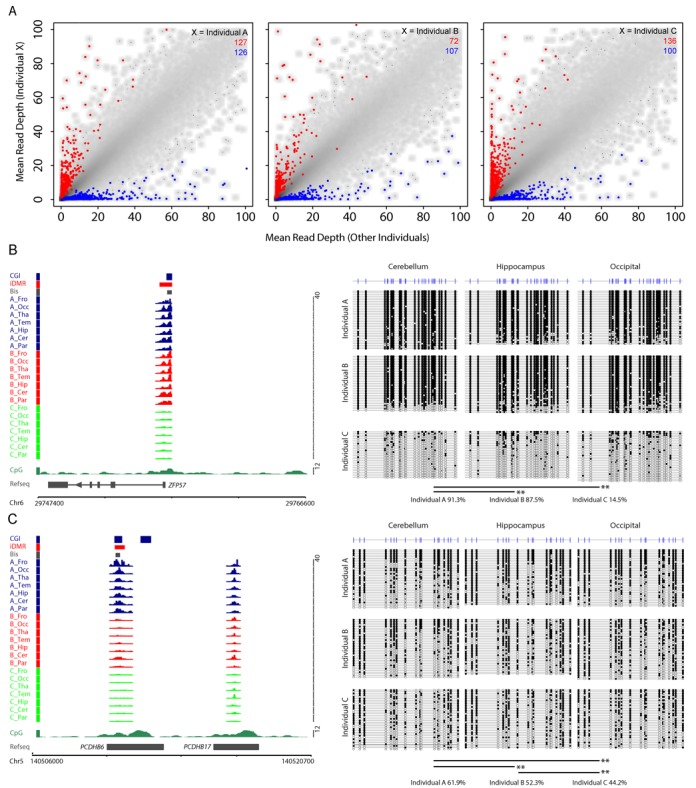
Identification of individual-specific DMRs (inDMRs). (A) Scatterplots depicting the averaged read depths for each individual versus the mean of the other two individuals. Individual-specific hyper- and hypomethylated DMRs are indicated for each individual (red and blue points, respectively). The number of hypermethylated (red) and hypomethylated (blue) DMRs are indicated for each comparison. (B) The left panel depicts the MAP-seq profiles for each individual (Individual A, darkblue; Individual B, red; Individual C, orange) at the ZFP57 locus (chr6 29746600–29766600; hg18 genome build). The right panel shows the bisulfite sequencing result confirming that Individual C has reduced methylation levels relative to Individuals A and B as predicted by the MAP-seq analysis. (C) The left panel depicts the MAP-seq profiles for each individual (Individual A, darkblue; Individual B, red; Individual C, orange) at the PCDHB6 locus (chr5 140505700–140520700; hg18 genome build). The right panel shows the bisulfite sequencing result confirming that the methylation level is the highest in Individual A and lowest in C as predicted by the MAP-seq analysis. The layout for (B) and (C) are presented as for Figure 2B.
