Figure 2.
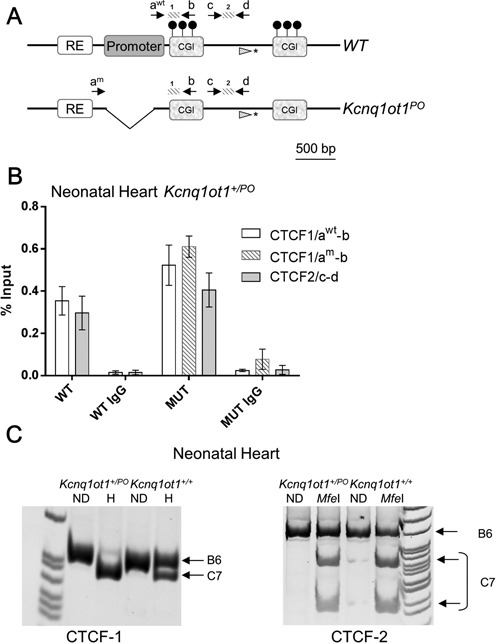
Chromatin immunoprecipitation analysis for CTCF at the KvDMR. (A) Schematic of the wild-type (WT) and Kcnq1ot1PO alleles. Primers awt and b amplify CTCF binding site 1 (CTCF-1) in the WT allele only, since awt anneals to a sequence in the promoter, which is absent in the mutant allele; primers am and b amplify CTCF binding site 1 in the mutant allele (PO). Primers c and d amplify CTCF binding site 2 (CTCF-2) of both the WT and Kcnq1otPO allele. Asterisk denotes SNP analyzed with the primer depicted with the hatched arrowhead. (B) Quantitative PCR of immunoprecipitated substrates from neonatal hearts of WT mice and Kcnq1ot1+/PO littermates. Assays were performed on mice from three litters each, in triplicate, and results expressed as percentage input normalized to H19 R1 (repeat 1 of the differentially methylated domain). Error bars indicate SEM. CTCF-1 shows occupancy of CTCF in both parental alleles of in both the WT mouse and the Kcnq1ot1+/PO mouse. For the Kcnq1ot1+/PO mouse (MUT), the white bar represents the data for the WT maternal (primers awt and b) and the hatched bar represents the mutant paternal allele (primers am and b) CTCF-2 shows occupancy of CTCF for both WT and mutant alleles. (C) Allele-specific restriction digests of ChIP-PCR products for primer pairs for CTCF-1 (left panel) and CTCF-2 (right panel). ND, non-digested; H, Hpy1881 digest. For CTCF-1, Hpy1881 cuts only the castaneus allele; for CTCF-2, MfeI cuts only the castaneus allele.
