Figure 2.
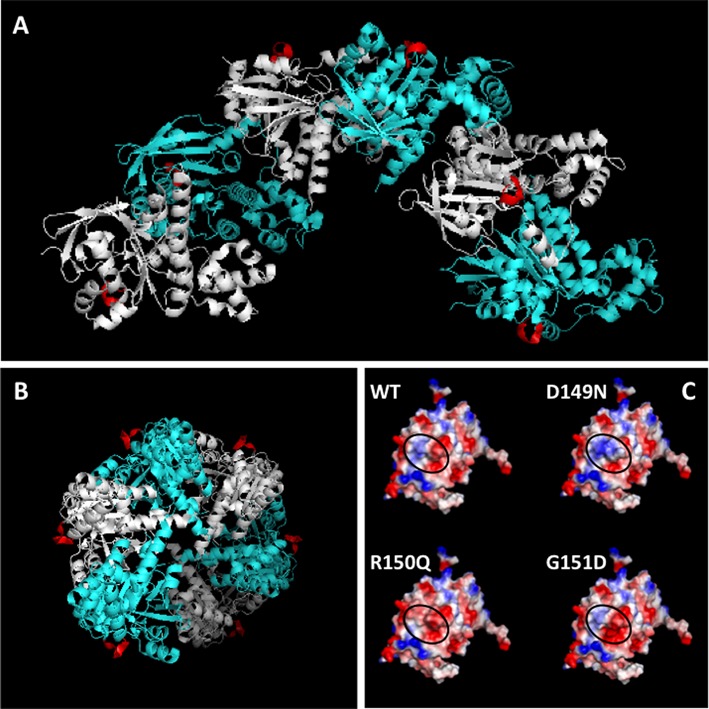
Location of the Schellman loop motif on the yeast Rad51 filament. (A) Structure of six contiguous subunits (one helical turn) of the Saccharomyces cerevisiae Rad51 filament (PDB ID: 3LDA), viewed perpendicular to the filament screw axis, with alternating subunits shown in cyan and silver. The Schellman loop motif on each subunit is highlighted in red. (B) End view, looking down the filament screw axis, which is perpendicular to the plane of the page. Color scheme as in (A), above. (C) Structure of the WT RAD51 core domain from PDB ID: 1N0W was altered to D149N, R150Q or G151D, and the resulting surface potentials were calculated. Blue denotes electropositive character, red denotes electronegative character and white denotes neutral character. The WT and variant proteins are shown in the same orientation. In each case, the black oval indicates the region of the protein surface corresponding to the Schellman loop.
