Figure 5.
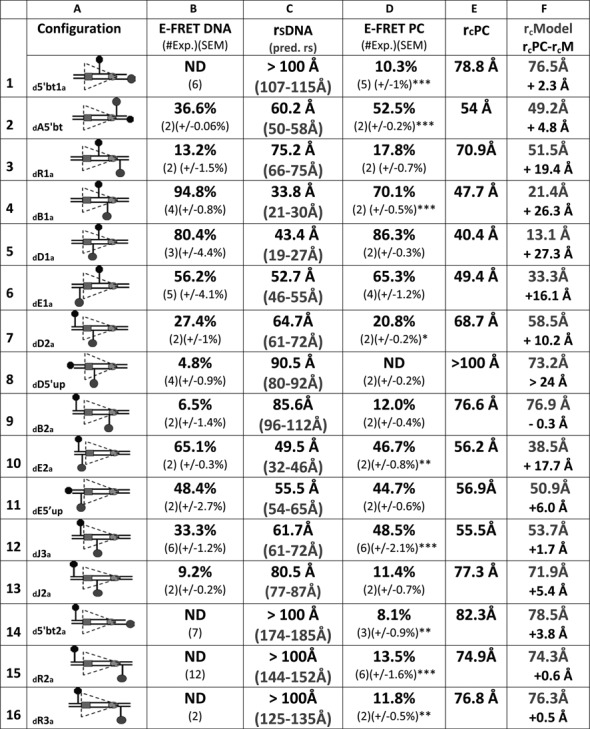
FRET analysis of the 12RSS in the PC. Data for 16 fluorophore-labeled substrates under PC conditions; in each panel, the columns designate the following features: column (A), schematic depiction of the substrates as in Figure 2; column (B), average energy transfer efficiency for substrate in the absence of protein (E-FRET DNA), with the number of independent experiments and SEM in parentheses (ND, no energy transfer above background); column (C), calculated distance between the donor and acceptor fluorophores in the substrate in the absence of protein (rsDNA) based on the E-FRET in column (B), with the predicted interfluorophore distance in the DNA (pred. rs) in gray in parentheses; column (D), average energy transfer efficiency for substrate in the complete reaction E-FRET PC, with the number of independent determinations and SEM in parentheses. Statistical comparison of E-FRET PC versus E-FRET DNA: *P < 0.05; **P < 0.01; ***P < 0.001; column (E), calculated distance between the donor and acceptor fluorophores in the substrate in the PC reaction rcPC, based on the E-FRET in column (D); column (F), interfluorophore distances in the PC models shown in Figure 7 of the 12RSS (rcModel, gray), estimated by measuring the distance between the 5′ carbons of the sugars of the two fluorophore-labeled nucleotides. Below (black), interfluorophore distance derived experimentally minus the interfluorophore distance in the model for the PC (rcPC-rcM).
