Figure 8.
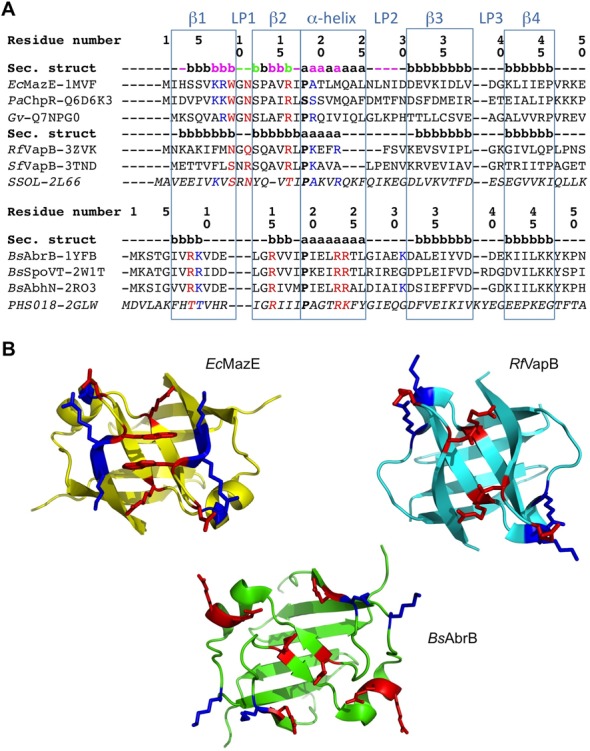
Structure-based sequence alignment of AbrB-like domain superfamily members. (A) Sequence alignment of the superfamily divided into two main families; the first one contains two subgroups. The consensus secondary structure within the superfamily is highlighted in light blue squares. Secondary structure elements within each family and sub-group are also given, representing the first one in each. The one belonging to EcMazE is color-coded by the CSP given in Figure 5 (Δδ > Δδav + 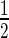 SD colored magenta, in green the residues of which their peaks disappear upon addition of the DNA). Residue numbering for the two families corresponds to that of EcMazE and BsAbrB, respectively. Key residues for DNA interaction are colored red, additional residues stabilizing the protein–DNA interaction in blue. The sequences from top to bottom are (corresponding PDB or Uniprot entries are given between parenthesis): EcMazE: Escherichia coli MazE antitoxin (1MVF); PaChpR: Pectobacterium atrosepticum ChpR suppressor of growth inhibitor (Q6D6K3); Gv: Gloeobacter violaceus cell growth regulatory protein (Q7NPG0); RfVapB: Rickettsia felis VapB antitoxin (3ZVK); SfVapB: Shigella flexneri VapB antitoxin (3TND); SSOL: Sulfolobus solfataricus transcription regulator (2L66); BsAbrB: Bacillus subtilis AbrB transition state regulator (1YFB); BsSpoVT: Bacillus subtilis SpoVT stage V sporulation protein T (2W1T); BsAbhN: Bacillus subtilis AbhN putative transition state regulator (2RO3); PHS018: Pyrococcus horikoshii S018 putative uncharacterized protein (2GLW). (B) Representative structures of the two AbrB-like domain families. Structures were superimposed using Pymol and thus are in the same orientation. Residues important for DNA binding are given in sticks and colored as defined in (A).
SD colored magenta, in green the residues of which their peaks disappear upon addition of the DNA). Residue numbering for the two families corresponds to that of EcMazE and BsAbrB, respectively. Key residues for DNA interaction are colored red, additional residues stabilizing the protein–DNA interaction in blue. The sequences from top to bottom are (corresponding PDB or Uniprot entries are given between parenthesis): EcMazE: Escherichia coli MazE antitoxin (1MVF); PaChpR: Pectobacterium atrosepticum ChpR suppressor of growth inhibitor (Q6D6K3); Gv: Gloeobacter violaceus cell growth regulatory protein (Q7NPG0); RfVapB: Rickettsia felis VapB antitoxin (3ZVK); SfVapB: Shigella flexneri VapB antitoxin (3TND); SSOL: Sulfolobus solfataricus transcription regulator (2L66); BsAbrB: Bacillus subtilis AbrB transition state regulator (1YFB); BsSpoVT: Bacillus subtilis SpoVT stage V sporulation protein T (2W1T); BsAbhN: Bacillus subtilis AbhN putative transition state regulator (2RO3); PHS018: Pyrococcus horikoshii S018 putative uncharacterized protein (2GLW). (B) Representative structures of the two AbrB-like domain families. Structures were superimposed using Pymol and thus are in the same orientation. Residues important for DNA binding are given in sticks and colored as defined in (A).
