Figure 3.
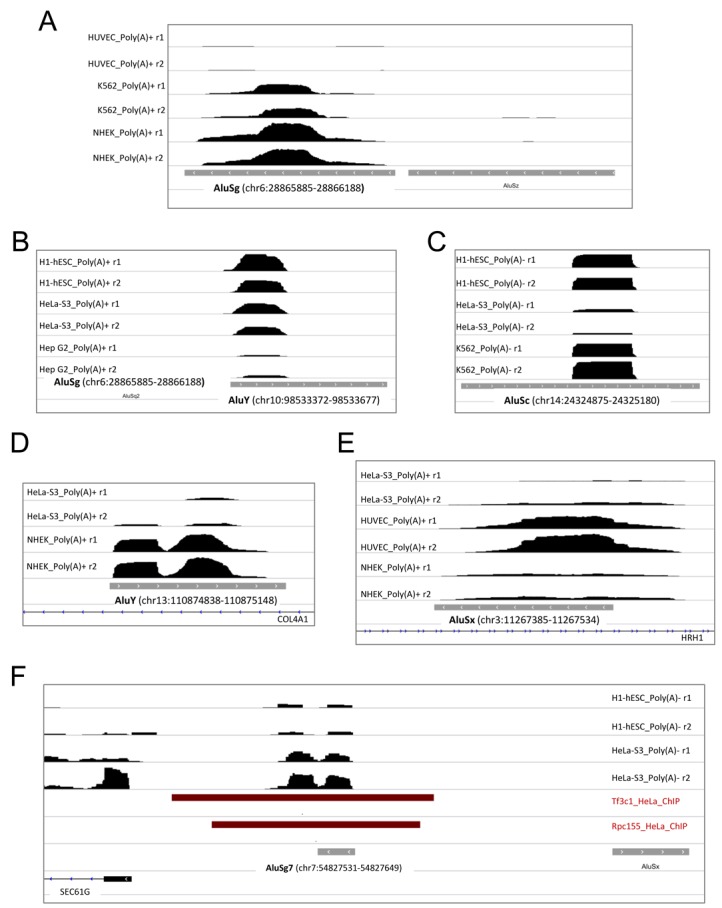
Base-resolution expression profiles for six representative Alus of the intergenic/antisense type. Panels A–C and F refer to purely intergenic Alus, panels D and E to two antisense Alus. Shown are the Integrative Genomics Viewer (IGV; http://www.broadinstitute.org/igv/home) visualizations of RNA-seq stranded expression profiles (in bigwig format) around Alu loci in the cell lines indicated either on the left (A–E) or on the right (F) of each panel. r1 and r2 indicate the two independent replicates found in ENCODE data. The orientation and chromosomal coordinates of each Alu, as well as the overlapping (antisense) or nearby RefSeq genes, are indicated in each panel. The dark red bars in panel F indicate regions associated to either TFIIIC (Tf3c1 track) or Pol III (Rpc155 track) in HeLa cells as derived from ENCODE ChIP-seq data.
