Figure 2.
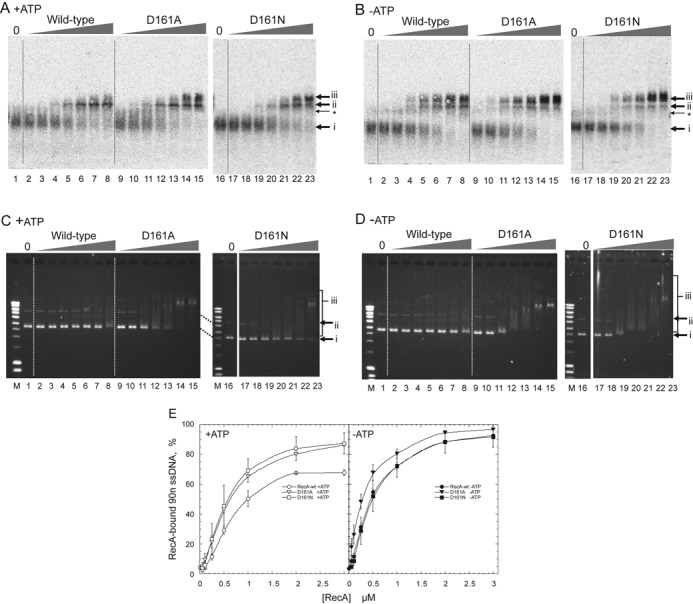
Amino-acid replacement of Asp-161 by Ala (D161A) or Asn (D161N) in loop L1 enhances double-stranded DNA-binding, but not single-stranded DNA-binding. (A, B) Single-stranded DNA-binding. 90-nucleotide single-stranded [33P]DNA (0.5 μM) was incubated with the indicated amounts of mutant or wild-type RecA for 5 min in the presence (A) or absence (B) of ATP in the standard reaction buffer for homologous joint formation. The samples were then fixed and subjected to agarose gel electrophoresis. The radioactivity in the gel was analyzed. Amounts of RecA in (A) and (B): lanes 1 and 16, no RecA; lanes 2, 9 and 17, 0.05 μM; lanes 3, 10 and 18, 0.1 μM; lanes 4, 11 and 19, 0.25 μM; lanes 5, 12 and 20, 0.5 μM; lanes 6, 13 and 21, 1.0 μM; lanes 7, 14 and 22, 2.0 μM; lanes 8, 15 and 23, 3.0 μM. Arrows i, unbound single-stranded DNA; ii and iii, RecA-bound single-stranded DNA. *Two single-stranded DNA-molecules that would interact with each other by local base-pairing. (C, D) Double-stranded DNA-binding. Unlabeled pBluescript SK(-) supercoiled (form I) double-stranded DNA (18 μM) was incubated for 15 min with the indicated amounts of mutant or wild-type RecA in the presence (C) or absence (D) of ATP in the standard reaction buffer for homologous joint formation. The samples were then subjected to agarose gel electrophoresis without fixation. The gel was stained with ethidium bromide and the DNA signals were detected by UV irradiation. Contrast was enhanced. Amounts of RecA: lanes 1 and 16, no RecA; lanes 2, 9 and 17, 0.5 μM; lanes 3, 10 and 18, 1.0 μM; lanes 4, 11 and 19, 2.0 μM; lanes 5, 12 and 20, 3.0 μM; lanes 6, 13 and 21, 4.0 μM; lanes 7, 14 and 22, 5.0 μM; lanes 8, 15 and 23, 10 μM. Lane M: DNA size markers. Arrows i, free form I; ii, nicked circular double-stranded DNA present in the form I preparation; iii, RecA-bound double-stranded DNA. (E) Quantitative representation of single-stranded DNA-binding analysis. The unbound 90-nucleotide single-stranded DNA among the total DNA signals in each lane was quantified, and the value of 100 minus% unbound single-stranded DNA was plotted as% RecA-bound single-stranded DNA (90n ssDNA) against the amounts of RecA. Each point indicates the average obtained from three independent experiments. ▽ (open inverse triangles), ▼ (closed inverse triangles), recA-D161A; ◻ (open square), ◼ (closed square), recA-D161N; ○ (open circles), ● (closed circles), RecA-wt. Open symbols, with ATP; closed symbols, without ATP.
