Figure 5.
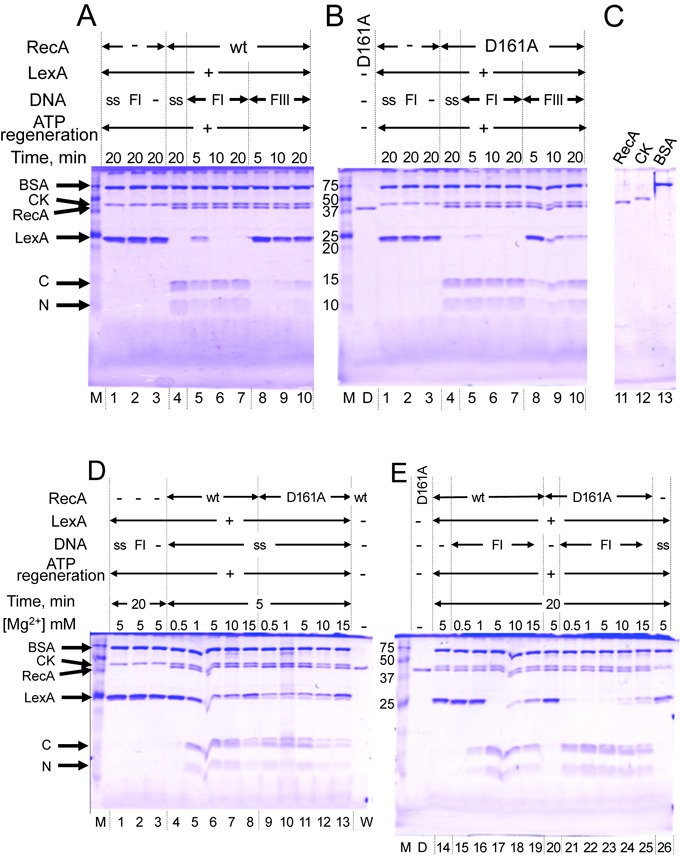
D161A replacement in loop L1 allows RecA to use double-stranded DNA as a cofactor for LexA repressor cleavage. (A, B, C) DNA cofactors for LexA cleavage. LexA (4.7 μM) was incubated for the indicated times with 1.0-μM RecA-wt (A) or recA-D161A (B) in the presence of the indicated DNA (18 μM) and 5.0-mM MgCl2. (D, E) Effects of MgCl2 concentrations on LexA cleavage. LexA was incubated for 5 or 20 min (as indicated) with 1.0-μM RecA-wt or recA-D161A in the presence of 18-μM single-stranded DNA (D) or double-stranded DNA (E), in reaction mixtures containing the indicated concentrations of MgCl2. DNA added to the reaction mixture: ss, single-stranded DNA; FI, negatively supercoiled closed circular DNA (form I DNA); FIII, linear double-stranded DNA (form III DNA). Proteins in signals were indicated on the left side of the panels: BSA, bovine serum albumin; CK, creatine phosphokinase from the ATP-regenerating system; RecA, RecA-wt or recA-D161A; LexA, uncleaved LexA; C and N, C-terminal and N-terminal fragments, respectively, of cleaved LexA. Lane M: protein markers, with sizes in kDa indicated on the left sides of the gels in (B) and (E); lane D, recA-D161A; lane W, RecA-wt.
