Figure 1.
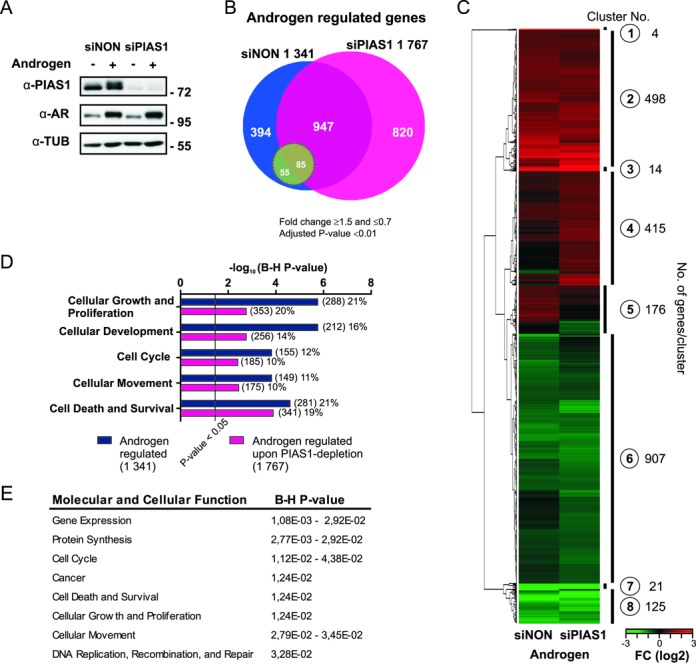
PIAS1 depletion affects androgen-regulated transcriptome of VCaP cells. VCaP cells were transfected with control siRNA (siNON) or PIAS1 siRNA (siPIAS1) and the cells were exposed to androgen (testosterone) or vehicle (ethanol) for 16 h, RNAs were isolated and microarray analysis was performed. (A) PIAS1 and AR protein levels in PIAS1-depleted VCaP cells. Cell lysates were immunoblotted with indicated antibodies. (B) Venn diagram showing androgen-regulated genes in siNON and siPIAS1 cells (1341 and 1767 genes, respectively). Green circle indicates AR-regulated genes significantly affected by PIAS1 depletion (when comparing fold changes of androgen-regulated genes in siNON- and siPIAS1-treated cells). (C) Heat map of the normalized expression values of differentially expressed transcripts in the BeadChip arrays based on unsupervised hierarchical clustering. (D) Biological processes of androgen-regulated transcripts in siNON and siPIAS1 cells as revealed by Ingenuity pathway analysis. Numbers in parentheses indicate the amount of transcripts linked to each biological process and percentages their relative portions among the androgen-regulated genes. Benjamini–Hochberg (B–H) multiple testing correlation was used for enrichment analysis. (E) Biological processes of the genes masked from androgen regulation by PIAS1 (820 genes) as revealed by IPA. B-H multiple testing correlation was used for enrichment analysis.
