Figure 1.
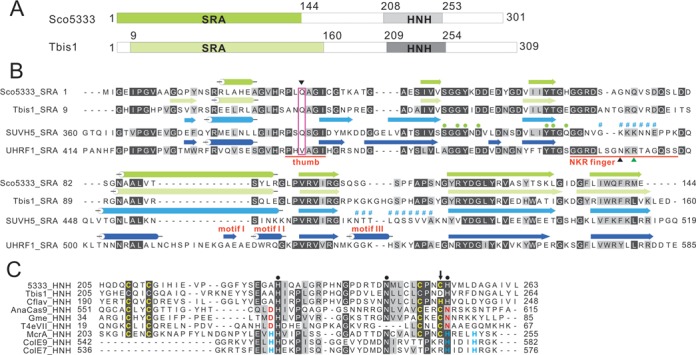
Sequence features of Sco5333 and Tbis1. (A) Domain architecture of Sco5333 and Tbis1, both of which contain a SRA domain (green) associated with a HNH motif (gray) at downstream. (B) Multiple sequence alignment and secondary structure comparison of SRA domains of Sco5333, Tbis1, SUVH5 and UHRF1. α-helices are in cylinders, and β-strands are in arrows. The thumb and NKR finger corresponding to UHRF1 are underlined in red. The inverted black triangle indicates the residue that inserts into the duplex and displaces the 5mC in the SUVH5 SRA. Filled green circles represent residues that interact with 5mC in the binding pocket of SUVH5 SRA and UHRF1 SRA, and the disordered region of SUVH5 SRA domain is represented by # #. Black and green upright triangles designate residues that replace the looped-out 5mC and mask the unmodified C in the UHRF1 SRA, respectively. (C) Multiple sequence alignment of HNH motifs of Sco5333, Tbis1, Cflav and the well studied HNH motif structure of AnaCas9, Gme, T4eVII, McrA, ColE9 and ColE7. Residues in golden designate the CCCC or CCCH type of zinc finger motifs. Filled black circles represent the conserved HNH motifs. The downward black arrow indicates that Tbis1 HNH displays a ruined CCCD zinc finger. The brown D and N are active sites of HNH motifs in AnaCas9, Gme and T4eVII that bind to divalent ions. The H in cyan designates the divalent ion-binding residues of HNH motifs in McrA, ColE9 and ColE7.
