Figure 6.
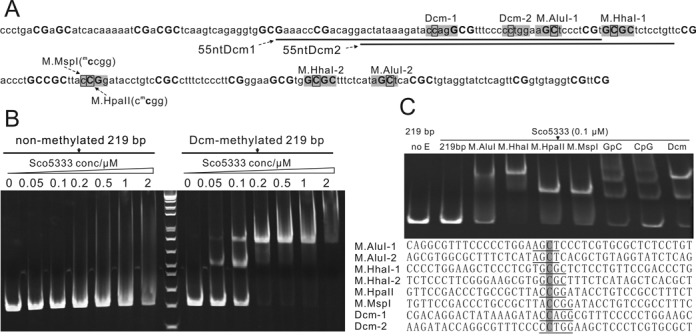
EMSA analysis of Sco5333 to methylated 219 bp derived from pUC18. (A) Sequence analysis of 219 bp which is PCR amplified from NT885–1103 of pUC18. Bases in shadow are modification sites of relative DNA MTases, and black rectangle designates the 5mC. C and G in bold represent the distribution of CpG and GpC modification sites. The underlined sequences are then synthesized and are named as 55ntDcm1 and 55ntDcm2. (B) Titration of Sco5333 on Dcm-modified and non-modified 219 bp fragment. Sco5333 shows high specificity to Dcm-modified 219 bp, and the two shifted bands in lane labelled 0.05 and 0.1 at right-half of the gel are due to the two Dcm modification sites which are centered at 219 bp. (C) EMSA of Sco5333 on 219 bp fragment modified by M.AluI, M.HhaI, M.HpaII, M.MspI, GpC, CpG or Dcm. Final concentration of Sco5333 is 0.1 μM. M.AluI, M.HhaI and Dcm have two modification sites on 219 bp, thus lanes 3, 4, and 9 show higher shift bands, whereas 219 bp modified by other MTases that have single modification site show lower shift bands. CpG and GpC have multiple modificaition sites (19 and 21 respectively), which result in multiple retarded bands. Statistics of Sco5333 binding sites on 219 bp shows poor sequence dependency.
