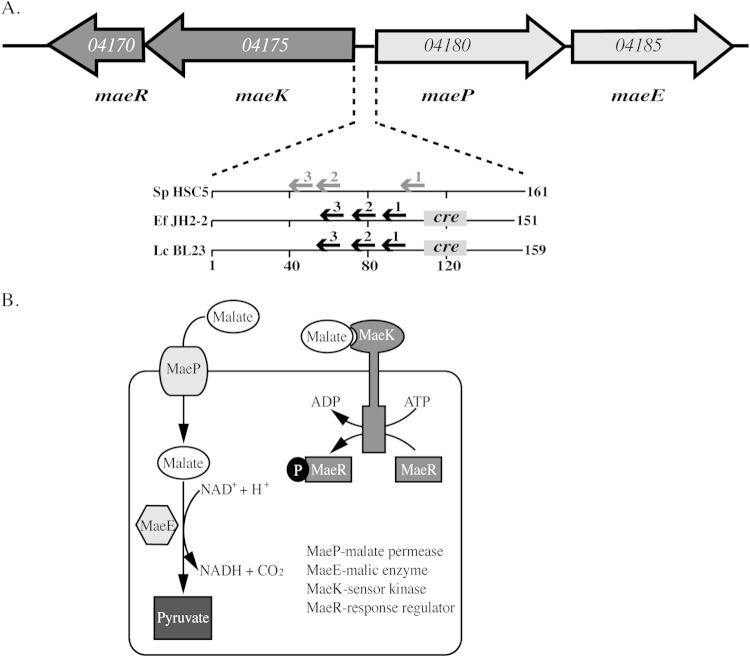
FIG 1.
The malic enzyme (ME) pathway in S. pyogenes. (A) The arrangement of the open reading frames that comprise the ME locus of S. pyogenes is shown by large arrows. Gene names are shown below, and the genomic loci listed within the open reading frames are based on the genome of S. pyogenes HSC5 (23). Known (black font) and predicted (gray font) regulatory elements of the intergenic region of S. pyogenes and Enterococcus faecalis JH2-2 (Ef JH2-2 [17]) and Lactobacillus casei BL23 (Lc BL23 [15]) are shown below. Arrows indicate sites in DNA bound by MaeR, while sites bound by CcpA are boxes labeled “cre” (catabolite regulatory element). Numbers below indicate intergenic distances in numbers of base pairs. (B) Schematic of the ME pathway. The subcellular localization, function, and reactions catalyzed by the various components of the ME pathway that are listed in the figure are shown.