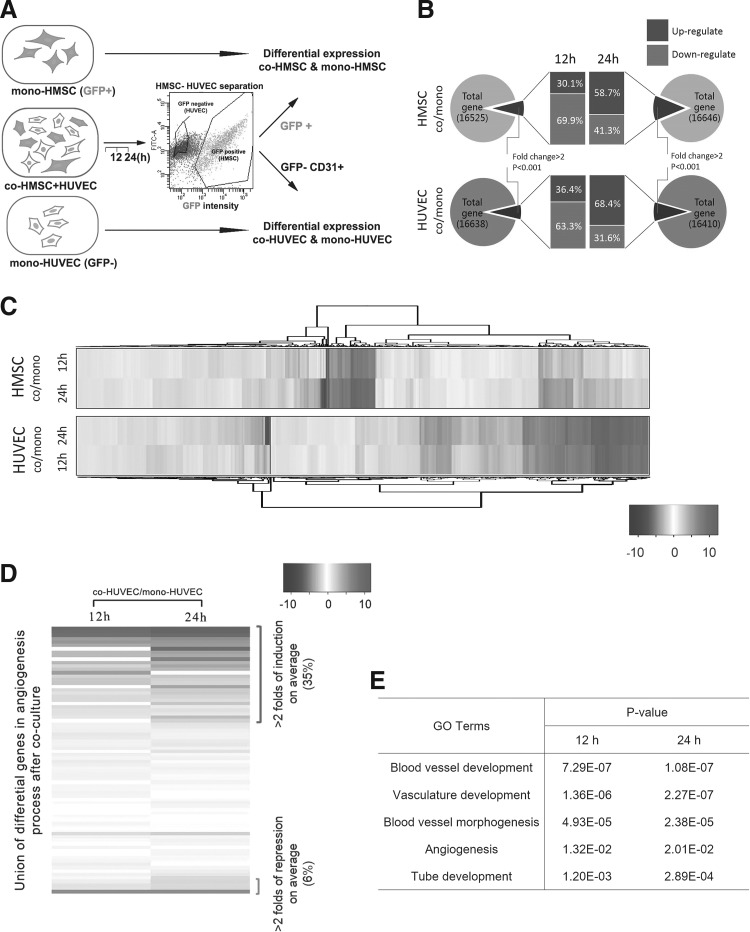
FIG. 2.
Coculture dynamically regulated the transcriptome alteration in both MSCs and HUVECs and raised angiogenic gene expression in HUVECs. (A) Schematic of FACS-based cell sorting after coculture and RNA-seq analysis. Co-HMSC was sorted based on GFP+ and co-HUVEC was sorted based on GFP− and CD31+. Transcriptome alteration was compared between cocultured MSC/HUVEC and their monoculture counterparts. (B) Statistical analysis of gene differential expression after coculture for 12 and 24 h separately in HMSCs and HUVECs. (C) Hierarchical clustering of RNA-seq data within genes both significantly altered in 12 and 24 h coculture for HMSCs and HUVECs. (D) Significant proportion of angiogenic related genes were elevated upon coculture in HUVECs. Heatmap was ordered via the fold of induction in Log2 scale. Significantly induced genes and repressed genes were indicated. (E) Gene ontology (GO) analysis on vascularization and angiogenesis-related process. Correlation of listed relevant process in 12 and 24 h coculture in HUVECs was evaluated by P value. FACS, fluorescence activated cell sortor; MSC, mesenchymal stem cell.