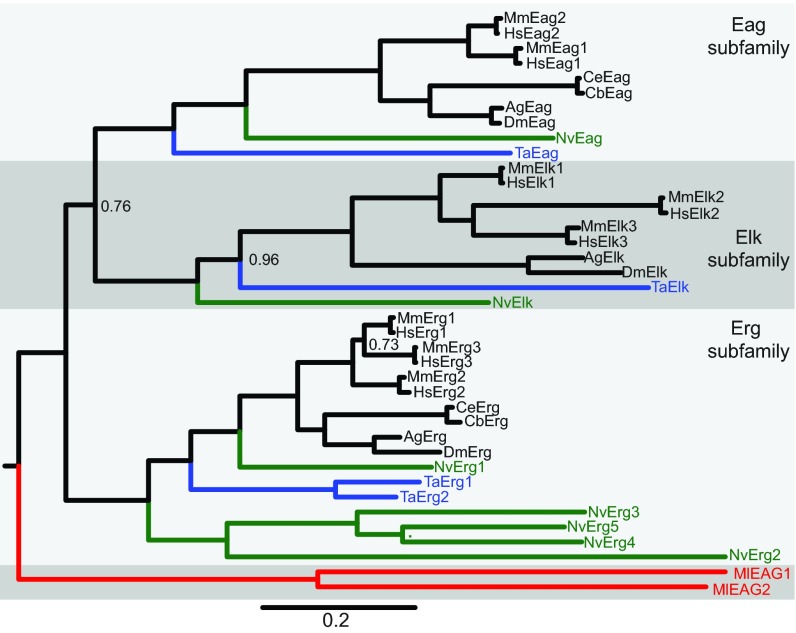
Fig. 1.
Phylogeny of the metazoan EAG K+ channel family. A Bayesian inference phylogeny of the EAG K+ channel family including sequences from bilaterians (black), cnidarians (green), placozoans (blue) and ctenophores (red). The phylogeny is midpoint rooted for display and the Eag, Elk and Erg family branches are labeled and differentially shaded. Ctenophore EAG channels branch separately from the Eag, Elk and Erg subfamilies, whereas cnidarian and placozoan EAG channels are placed within the subfamilies. The scale bar indicates substitutions/site and node posterior probabilities of less than 1 are indicated. Most nodes had a posterior probability of 1, indicating a strong support for the phylogeny. All sequences used in the phylogeny are provided in supplementary material Table S2, with database links to sequence resources. Species prefixes are as follows: Ag, Anopheles gambiae (mosquito); Cb, Caenorhabditis briggsae (nematode); Ce, Caenorhabditis elegans (nematode); Dm, Drosophila melanogaster (fruit fly); Hs, Homo sapiens (human); Ml, Mnemiopsis leidyi (ctenophore); Mm, Mus musculus (mouse); Nv, Nematostella vectensis (sea anemone); Ta, Trichoplax adhaerens (placozoan).