Abstract
The human Mediator complex is a central integrator for transcription and represents a primary interface that allows DNA-binding transcription factors to communicate their regulatory signals to the RNA polymerase II enzyme. Because Mediator is dynamic both in terms of subunit composition and structure, it presents challenges as a target for small molecule probes. Moreover, little high-resolution structural information exists for Mediator. Its global requirement for transcription, as well as its distinct, transcription factor specific interaction surfaces, however, suggest that development of probes that bind specific Mediator subunits might enable gene- and pathway-specific modulation of transcription. Here we provide a brief overview of the Mediator complex, highlighting biological and structural features that make it an attractive target for molecular probes. We then outline several chemical strategies that might be effective for targeting the complex.
Keywords: gene expression, CDK8 module, Mediator complex, probe discovery
1. Introduction
The Mediator complex is a large, multi-subunit assembly that is generally required for transcription.[1] Mediator exists primarily in two different forms: a “core” complex and a “CDK8-Mediator” complex. In human cells, the core Mediator complex consists of 26 subunits, whereas the CDK8-Mediator complex consists of 29 subunits (25 of which are shared with core Mediator). Although evolutionarily conserved from yeast to human, Mediator has evolved rapidly, acquiring metazoan-specific subunits and new biological functions. Known links between Mediator subunits and human disease are many, and are increasing each year (reviewed in [2] and [3]). For these and other reasons, Mediator is an attractive, albeit challenging, target for molecular probes.
2. A central role for Mediator as a general and gene-selective regulator of transcription
The regulation of gene expression is fundamental for every major physiological process, and changes in gene expression patterns are hallmarks of human development, disease, and adaptive responses (e.g. viral infection). Many well-studied signaling cascades that initiate in response to environmental cues ultimately trigger changes in gene expression.[4] Activation (and repression) of pathway- and stimulus-specific genes is achieved in large part by DNA-binding transcription factors (TFs), which serve as the primary drivers of cell state and cell physiology.[5] As a testament to their enormous regulatory power, an entire population of fibroblasts or adipocytes can be converted to myotubes upon expression of a single TF, MyoD.[6, 7] Given the cornerstone roles of TFs in controlling human development, physiology, and disease, an effective strategy to manipulate TF function would have widespread impact on human health.[8,9] The traditional (and reasonable) concept that TF activity is best manipulated by directly targeting the TF itself has not yielded much progress.[10,11] We outline here a different approach that circumvents the TF entirely, yet has potential to achieve some of the same goals.
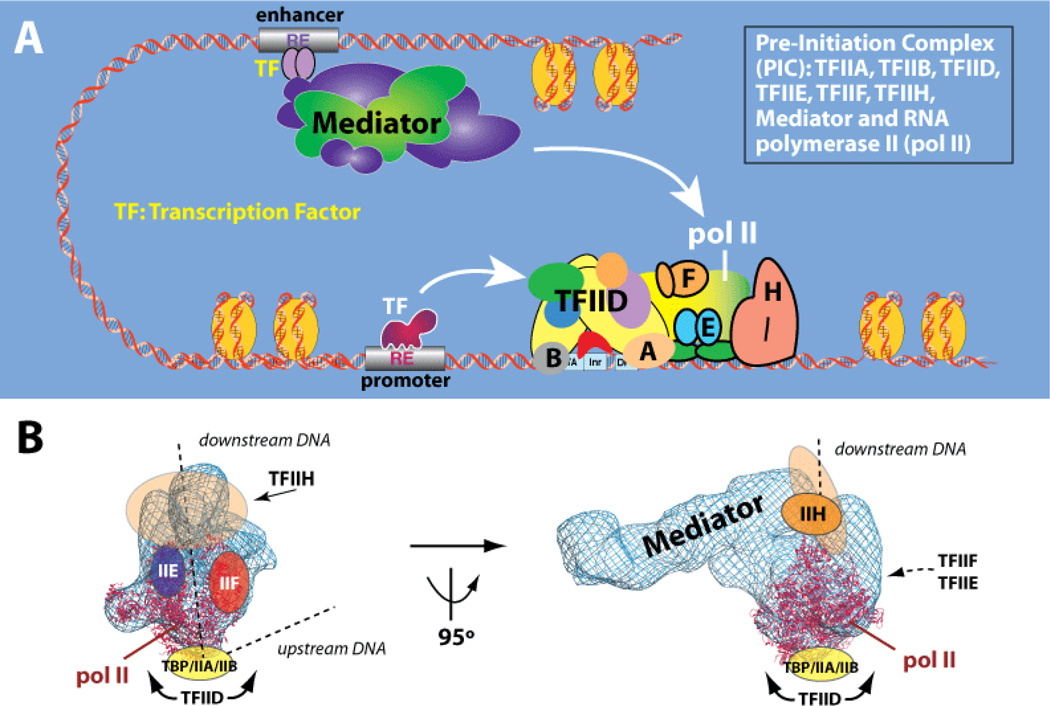
It is widely understood that sequence-specific DNA-binding TFs activate or repress transcription by somehow affecting RNA polymerase II (pol II) activity. The pol II enzyme is responsible for transcription of all protein-coding and most non-coding RNA genes. Remarkably, TFs do not bind pol II directly and instead bind the large, multi-subunit Mediator complex.[12,13] Mediator, in turn, interfaces extensively with pol II and relays regulatory signals from TFs directly to the pol II enzyme (Figure 1A, B). Current understanding suggests that Mediator and the large, multi-subunit TFIID complex—not pol II—represent the key factors through which TFs regulate transcription. In fact, the Mediator complex appears to be as important to controlling expression of protein-coding genes as the pol II enzyme itself.[14,15] In agreement with this, Mediator has been described as the endpoint of cell signaling pathways because it represents the ultimate, functional target for DNA-binding TFs.[16,17]
Figure 1.
Mediator is the central scaffold around which the human transcription machinery assembles. (a) Schematic of the human “Pre-Initiation Complex” (PIC). Complexes are shown at same relative scale. (b) Structural model of the human PIC, based upon a cryo-EM study of a Mediator-pol II-TFIIF assembly.[18]
Gene expression (a.k.a. transcription) occurs within the context of a macromolecular assembly known as the Pre-Initiation Complex (PIC). The PIC consists of eight protein complexes, including Mediator and the pol II enzyme, and is approximately four MDa in size (Figure 1A). At 1.2 MDa, the Mediator complex represents a major sub-assembly within the PIC, and structural and functional studies have revealed that Mediator acts as a scaffold around which the human transcription machinery assembles (Figure 1B).[18–22] This central structural role for Mediator can at once explain its general requirement for transcription, and its ability to regulate the function of pol II at different stages of the transcription cycle, such as initiation and elongation.[23]
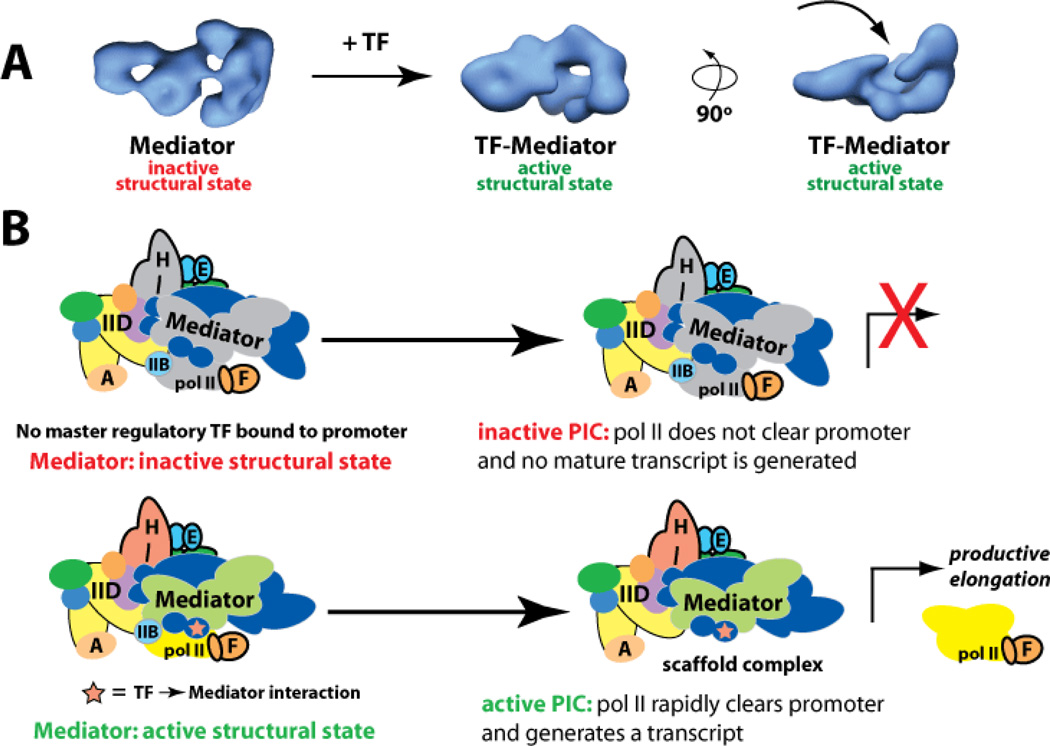
Mediator is targeted by a vast array of DNA-binding TFs[24] that regulate fundamental physiological responses and are implicated in myriad human diseases. Whereas multiple labs have established that TFs activate transcription upon binding the Mediator complex,[25–32], precisely how TFs control pol II function via the Mediator complex remains poorly understood. Data continue to suggest, however, that structural shifts in Mediator, triggered by TF binding, play an essential role.[33,34] For example, upon binding Mediator, the activation domain of the p53 TF induces conformational changes in Mediator that correlate with activation of pol II at the promoter (Figure 2). In the absence of a key p53→Mediator interaction, Mediator adopts alternate structures that appear to maintain pol II in an inactive state.[35] Thus, the TF→Mediator interface appears to represent a key “control point” that dictates TF function.
Figure 2.
Model for how TFs activate transcription via Mediator. (a) TFs induce sweeping structural shifts upon binding Mediator. At left is the structure of Mediator without a TF bound, whereas at right are two views of a TF-bound state (in this case, p53-Mediator). A common feature of TF-bound Mediator structures is a large pocket domain (arrow) that corresponds to the pol II binding site. (b) Schematic of an inactive or active PIC. In the absence of a key regulatory TF, the PIC can assemble at the promoter but remains in an inactive or unproductive structural state. Upon TF-Mediator binding, Mediator adopts an active structural state, which activates the PIC and pol II transcribes the gene. Adapted from ref. [35].
3. Targeting Mediator and Mediator-TF interactions with small molecules
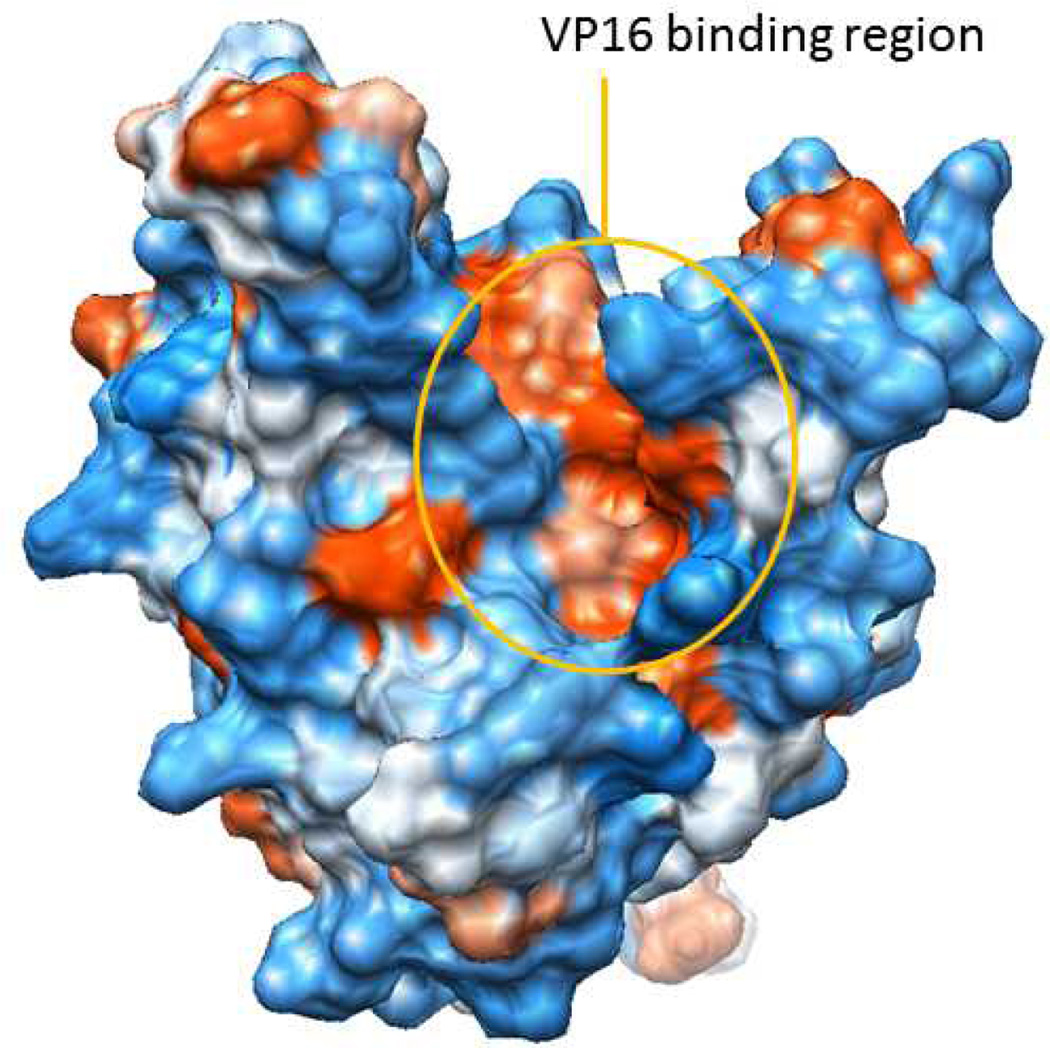
Like many multiprotein complexes, Mediator offers both challenges and opportunities for small molecule ligand discovery. Dogma dictates that proteins and protein-protein complexes without enzymatic function, or that do not have cognate small molecule ligands, largely fall into a realm of ‘undruggable’.[36] Much of the failure of high-throughput screening against protein-protein interaction (PPI) targets has been attributed to the inadequacies of current compound collections to recapitulate the physical features of PPIs.[37,38] However, contemporary structural studies are providing an initial round of insights into the interfaces between the various Mediator subunits that are encouraging. For example, several structural studies are now completed for Mediator-TF interfaces.[39] NMR studies have been successful in characterizing the interaction between the activation domain of SREBP with MED15 and VP16 with MED25.[40–42] A challenge, however, will be to develop a small molecule that can selectively block the Mediator-TF interface. Although only a limited number of such interfaces have been structurally characterized thus far, it appears that some will be more amenable to small molecule targeting than others. For example, whereas the SREBP-MED15 interaction surface is relatively compact,[40] NMR analyses of VP16-MED25 show evidence of multiple distinct bound conformations that involve opposite faces of MED25.[41,42] Despite this, a prominent MED25 hydrophobic cleft, formed from two α-helices and a β-barrel, was identified as a key region for VP16 binding (Figure 3). Mutations in the region abrogated VP16 binding[41] and provide some suggestions for small molecule antagonists that exploit the deep hydrophobic pocket that dominates this region.
Figure 3.
The VP16 binding region of MED25 involves a deep hydrophobic pocket at the intersection of two α-helices and a β-barrel.
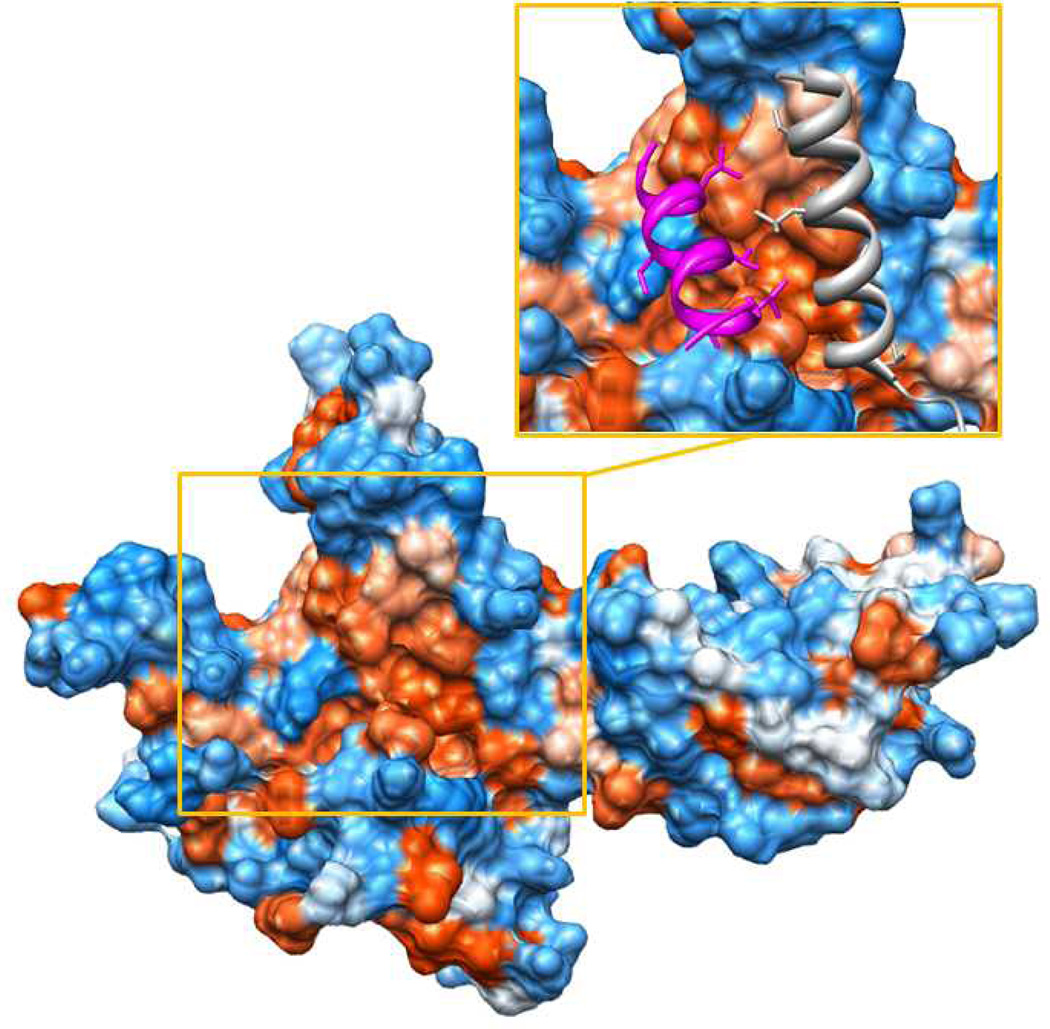
Beyond Mediator-TF interactions, it may be feasible to target subunit-subunit interactions within Mediator itself. In principle, this could achieve outcomes similar to blocking a specific Mediator-TF interaction. The structural integrity of Mediator can be maintained in the absence of select subunits,[17,43,44] indicating that Mediator still functions as a genome-wide regulator of transcription even if certain subunits are lacking. The absence of a particular Mediator subunit can preclude gene activation by its corresponding TF target. For example, knocking out MED1 or MED23 does not appear to greatly compromise Mediator structural stability, but resulted in gene-selective effects on transcription.[45] MED1 knockout murine embryonic fibroblasts were unable to appropriately activate nuclear receptor targets (which interact with MED1), whereas MED23 knockout murine embryonic stem cells could not activate select ELK1 target genes (ELK1 binds MED23).[17,46] High resolution structural data of Mediator subunit-subunit interfaces would facilitate identification of molecular interfaces to target with small molecules. Whereas these data are largely unavailable for human Mediator, a significant amount of data are available for yeast versions of the complex.[39] In fact, a majority of the 7-subunit “head module” of yeast Mediator (representing about 20% of the yeast Mediator complex) has been crystallized by several groups.[47–49] These data have identified numerous and extensive interfaces between subunits. As one example, X-ray crystallography of the MED17–MED11–MED22 subcomplex of the S. cerevisiae head domain of Mediator reveals that MED11 and MED22 interact with MED17 along a large, deep, hydrophobic cleft (Figure 4). Although speculative, this pocket may provide an appropriate interface for binding a small molecule. Whether structural features of subunit-subunit interfaces are conserved between yeast and human Mediator remains to be seen. Although the sequence conservation between yeast and human Mediator subunits is very poor, structural homology could still be preserved. In fact, crystal structures of the Mediator head module from two different yeast strains (S. cerevisiae vs. S. pombe) are highly conserved despite only 15% sequence identity.[48]
Figure 4.
MED17 interacts with MED11 and MED22 along a hydrophobic groove. Insert: Two alpha helices from MED11 (magenta) and MED 22 (silver) project hydrophobic residues into the MED17 hydrophobic groove.
The studies outlined above provide early structural information in support of the idea that the interactions between Mediator subunits or Mediator and TFs could be modulated by small molecules. Compounds that could block, or mimic, the functions that arise from TF→Mediator binding would provide powerful capabilities to affect transcriptional programs. However, such compounds would have limited use if every TF targeted the same surface on the Mediator complex. To a large extent, this is not the case for Mediator (Figure 5). In fact, unlike other transcription regulators that also bind TFs, such as CBP/p300,[50,51] distinct TFs target completely different surfaces on Mediator;[24,52] moreover, these distinct TF→Mediator interactions can induce different Mediator structural states, suggesting that Mediator adopts TF-specific functionality.[33,53] Taken together, these observations suggest a potential for selective regulation of TF function, and that targeting specific surfaces within Mediator (e.g. the SREBP binding site within MED15) may not impact gene expression patterns governed by other TFs.
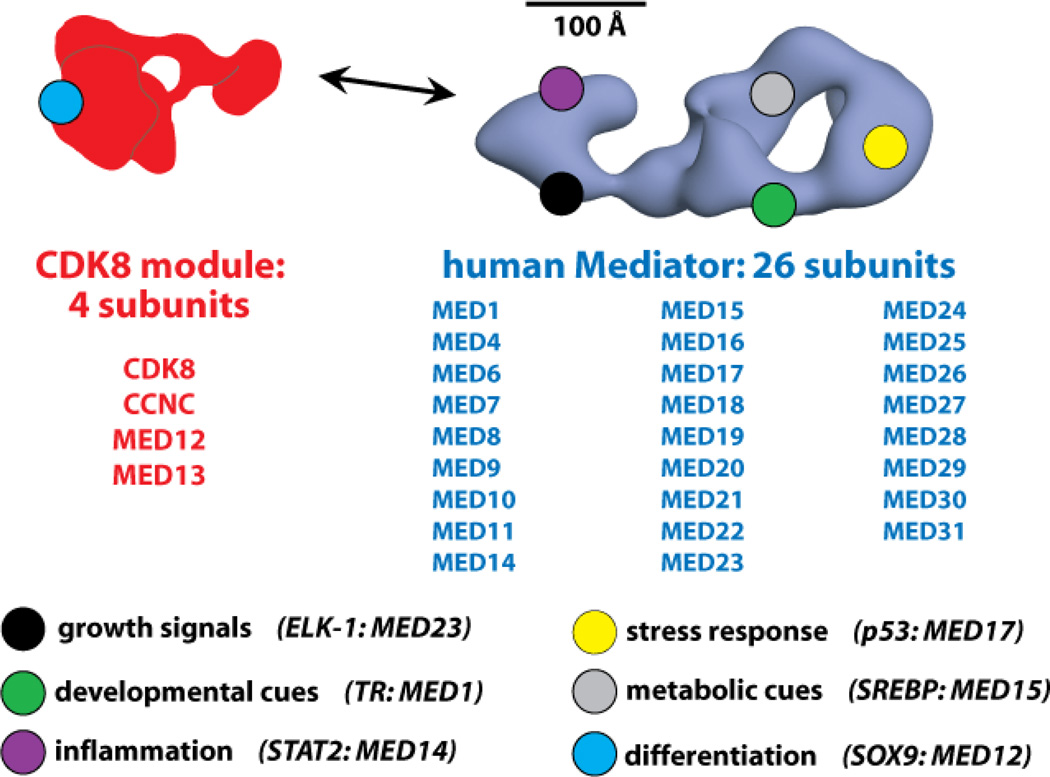
Figure 5.
Distinct TFs bind different Mediator subunits. The CDK8 module (red) and the Mediator complex (blue) are shown, along with the subunit composition of each. Because different TFs regulate distinct cellular processes, the varied TF-Mediator interaction surfaces can be considered key control points for these TF-directed events. Examples of basic physiological processes, and the TFs that help regulate them, are shown at the bottom. The Mediator subunit target of the TF is also listed. The general locations shown for select subunits are for illustrative purposes only; the location of each Mediator subunit is not well defined, especially for human Mediator.
Several “artificial transcription factors” have been developed that bind Mediator and activate transcription in reporter assays at artificial promoters.[54–58] Moreover, it has been demonstrated that TF-cofactor interactions can be selectively targeted and disrupted by small molecules.[59,60] These findings establish that 1) the Mediator complex can be successfully targeted with small molecules to impact gene expression, and 2) TF-cofactor interfaces (e.g. NOTCH transactivation complex) can be blocked with small molecules. These observations suggest it might be possible to develop small molecules that target key TF-Mediator interfaces to selectively regulate gene expression programs. Below, we outline three areas of contemporary chemistry that are impacting the discovery of molecules with function against protein-protein interfaces and that may offer avenues to modulation of Mediator function (Figure 6).
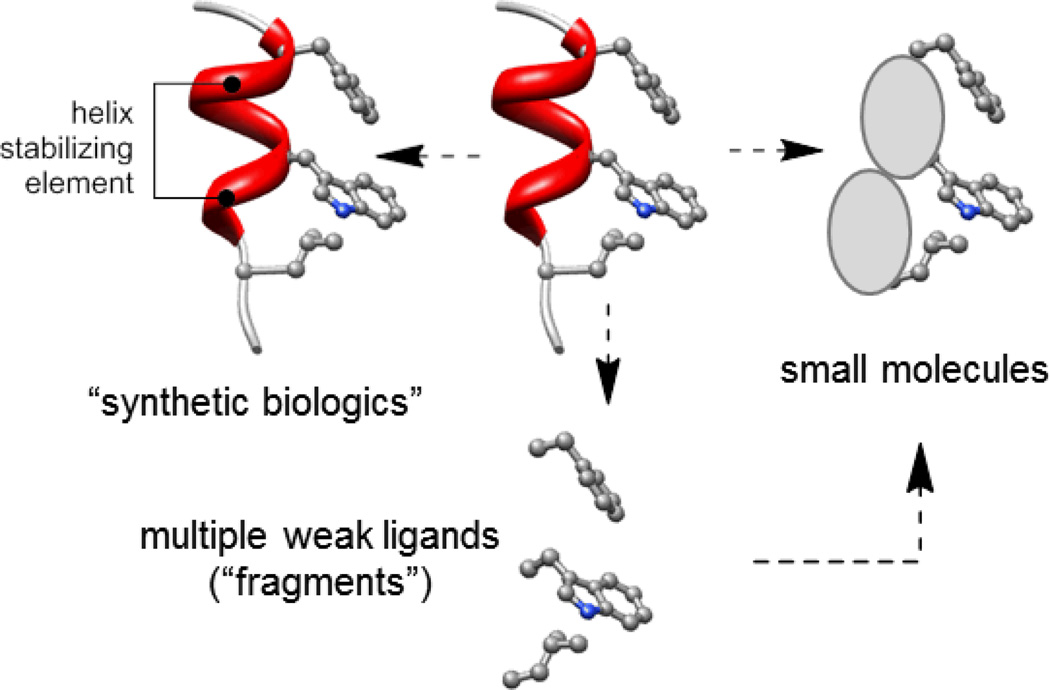
Figure 6.
Three areas of contemporary chemistry that are impacting the discovery of functional molecules against protein-protein interfaces: a. structural homologs e.g. stapled peptides and related compounds form a family that are increasingly being referred to as “synthetic biologics”; b. small molecules from diversity-oriented synthesis and other approaches; c. weak ligands identified by high-concentration screening (fragments) that are optimized to give potent small molecules.
3.1 Peptides, peptidomimetics, and stapled peptides
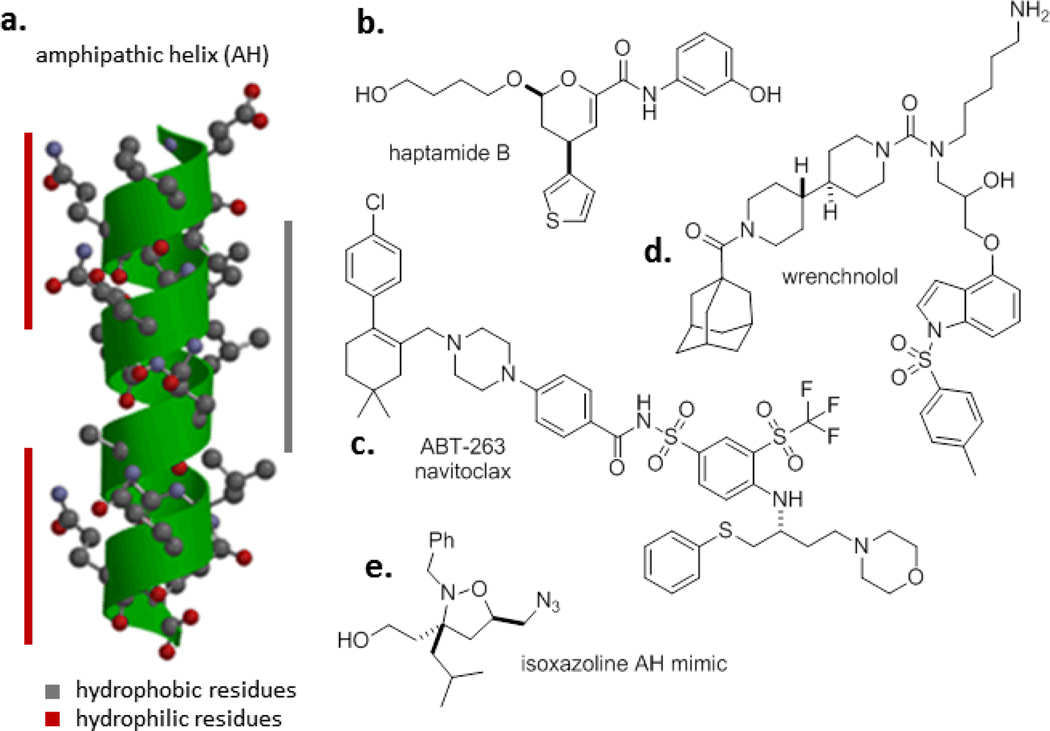
Although peptides that modulate the function of target systems can often be discovered by approaches that span synthesis through phage display, short peptides are often viewed as poor starting points for therapeutics. This is due to (i) sensitivity to proteolysis, (ii) generally poor transport properties, which can hinder applications against intracellular targets, and (iii) diminished structural organization when compared with larger domains.[61] Notwithstanding these caveats, short peptides and related peptoids, peptidomimetics, and stapled peptides have a proven track record in both abrogating and reconstituting PPIs.[62,63] In an early example relevant to control of transcription, fusing a 15-mer peptide (which was presumed to form an amphipathic helix that recruited coactivators) to the Gal4 DNA-binding domain resulted transcription at levels ~20% of the full-length Gal4 activator.[64] Functional small molecule peptidomimetics of these amphipathic helices based on isoxazolines have been developed, although their potential has yet to be widely explored.[65–67] Peptides that bind yeast Gal11 (Med15) are also functional in activating transcription, albeit at lower levels than the natural activation domain.[58]
The success of peptides against challenging PPI targets is driving efforts to improve their pharmacokinetic properties. Although general solutions to problems associated with cellular permeability have yet to emerge, binding affinities and selectivities can often be improved by the addition of elements that enhance secondary structure, such as lactam or disulfide bridges between appropriately placed side chains.[68,69] In addition, foldamers based on β-peptides often display higher propensities toward secondary structure than the corresponding native peptide.[70] Although these technologies have yet to be employed against PPIs in the Mediator complex, they have provided encouraging success against a number of complex PPI targets and the prevalence of helical recognition domains in transcriptional complexes suggests likely future prospects.[71] For example, disulfide-bridged 9-mer peptides that show increased helicity relative to acyclic structures inhibit the interaction between the estrogen receptor α (ERα) and a co-activator with a Ki of 25 nM.[72]
Among the most recent advances in this arena are hydrocarbon stapled peptides, which are often based on ring-closing metathesis strategies.[73,74] In cases where systems are well characterized structurally, this can be a fruitful path from structure to a functional molecule. Examples of success include the discovery of ligands that antagonize the TP53-HDM2 interaction,[75] the BID-BAX interaction,[76] and ER/co-activator interactions.[77] In several cases structures of the ‘stapled’ peptides bound to the target have shown faithful reproduction of major interactions found in the native peptide, as well as additional interactions between the hydrocarbon staple and the target.[78] In a particularly impressive example, a stapled peptide derived from the coactivator MAML1 was able to bind to the NOTCH1-transcription factor complex and modulate recruitment of MAML1.[59] Extensive studies connected the observed in vivo activity of this peptide with inhibition of the NOTCH pathway.
3.2 Small molecules, diversity-oriented synthesis, and microarrays
Collections of small molecules have played an important role in the rapid rise of high-throughput screening (HTS) as a ligand discovery paradigm. Screening libraries have a strong bias toward the chemical space that has proven successful against historically ‘druggable’ targets, and broadly speaking HTS produces low hit rates against PPI targets. Nonetheless, there have been successes against PPIs,[79] including the discovery of small molecule inhibitors of the interaction between the TF Elf3 and MED23 that were optimized to give wrenchnolol (Figure 7).[54,55] Subsequent studies involving a bivalent construct of wrenchnolol and a polyamide DNA-targeting construct produced robust activation of transcription both in vitro and in cells.[56]
Figure 7.
Examples of molecules active against targets in transcription and protein-protein interfaces.
Studies at the intersection of new chemical space and novel screening methods have also provided indications that TFs and protein complexes containing them can be directly modulated by small molecules stemming from diversity-oriented synthesis (DOS).[80,81] A small molecule microarray-based screen of 12,396 molecules identified DOS-derived hits that bound Hap3p (a subunit of the S. cervevisiae Hap2/3/4/5p TF complex) and modulated transcription. Optimization led to haptamide B, which binds Hap3p with a dissociation constant of 0.33 μM and reduces expression of Hap3p dependent genes in cells.82] Other examples of DOS-derived molecules that modulate PPIs and challenging targets in transcriptional biology include the function of Ure2p in glucose signaling in yeast,[83] and the discovery of lactam carboxamides that inhibit the interaction of the HOXA13 TF with DNA.[84]
3.3 Fragment-based methods
In the past decade, the application of strategies involving high-concentration screening or sophisticated structural methods as a mechanism to discover low-molecular weight and (often) weak-affinity ligands has grown in popularity.[85] This approach – often called ‘fragment-based ligand discovery’ – is predicated on the power of modern medicinal chemistry to evolve potent small molecule ligands when supported by structural information and has proven success against a variety of targets including PPIs.[86] In a flagship example, Abbott used NMR spectroscopy to discover weak ligands that inhibit the Bcl-xL/Bak protein-protein interaction.[87] The initial ligand discovered was evolved by the combination of synthesis and structural biology to provide several generations of compounds that culminated in ABT-263, which has low nM Ki values against Bcl-2 family proteins, and is also orally bioavailable.[88] The application of approaches to ligand discovery against the Mediator complex based on structural information and that begin with fragments seems likely to be fruitful given the increasing amount of structural information available on Mediator and its interactions.
4. Conclusions
The large size and conformational flexibility of Mediator provides an extensive surface for potential protein-protein interactions. The fact that many different TFs bind Mediator at distinct sites along its surface (i.e. via different Mediator subunits) suggests that targeting such sites could provide a measure of selectivity. Several groups have successfully targeted Mediator with small molecule “artificial transcription factors” that could forecast future progress with new classes of molecular probes. Significant challenges include: 1) the structural flexibility of Mediator, which could mask the target site in specific contexts. 2) For some interfaces, the TF-Mediator interaction may occur through multiple low-affinity sites that would be difficult to mimic with a small molecule. Although it is unclear how common this may be, an example of such an interaction has been characterized in yeast.[89–91] 3) Another challenge will be to develop compounds with sufficient binding affinity to modulate the action of the cognate TF. The few known human Mediator-TF binding affinities are in the nM range (VP16-MED25 and SREBP-MED15). Similarly, attention must be paid to the physical properties of ligands that will require cellular or nuclear uptake for action. Despite these challenges, the success of molecules such as ABT-263 provide much support to the idea that the modulation of complex biological functions is possible with small molecules.
References
- 1.Taatjes DJ. Trends Biochem Sci. 2010;35:315. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spaeth JM, Kim NH, Boyer TG. Seminars in cell & developmental biology. 2011;22:776. doi: 10.1016/j.semcdb.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Napoli C, Sessa M, Infante T, Casamassimi A. Biochimie. 2012;94:579. doi: 10.1016/j.biochi.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Cell. 2011;144:646. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Lee TI, Young RA. Cell. 2013;152:1237. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lassar AB, Thayer MJ, Overell RW, Weintraub H. Cell. 1989;58:659. doi: 10.1016/0092-8674(89)90101-3. [DOI] [PubMed] [Google Scholar]
- 7.Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. Science. 1988;242:405. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 8.Engelkamp D, van Heyningen V. Current opinion in genetics & development. 1996;6:334. doi: 10.1016/s0959-437x(96)80011-6. [DOI] [PubMed] [Google Scholar]
- 9.Jimenez-Sanchez G, Childs B, Valle D. Nature. 2001;409:853. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- 10.Darnell JE., Jr Nat Rev Cancer. 2002;2:740. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 11.Koehler AN. Curr Opin Chem Biol. 2010;14:331. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conaway RC, Conaway JW. Curr Opin Genet Dev. 2011;21:225. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik S, Roeder RG. Nat Rev Genet. 2010;11:761. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takagi Y, Kornberg RD. J Biol. Chem. 2006;281:80. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 15.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Cell. 1998;95:717. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 16.Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Proc. Natl. Acad. Sci. USA. 1998;95:8538. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens JL, Cantin GT, Wang G, Shevchenko A, Shevchenko A, Berk AJ. Science. 2002;296:755. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- 18.Bernecky C, Grob P, Ebmeier CC, Nogales E, Taatjes DJ. PLoS Biol. 2011;9:e1000603. doi: 10.1371/journal.pbio.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cantin GT, Stevens JL, Berk AJ. Proc Natl Acad Sci U S A. 2003;100:12003. doi: 10.1073/pnas.2035253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Black JC, Choi JE, Lombardo SR, Carey M. Mol Cell. 2006;23:809. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Chen XF, Lehmann L, Lin JJ, Vashisht A, Schmidt R, Ferrari R, Huang C, McKee R, Mosley A, Plath K, Kurdistani SK, Wohlschlegel J, Carey M. Cell reports. 2012;2:1061. doi: 10.1016/j.celrep.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson KM, Wang J, Smallwood A, Arayata C, Carey M. Genes & Development. 2002;16:1852. doi: 10.1101/gad.995702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conaway RC, Conaway JW. Biochim Biophys Acta. 2013;1829:69. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borggrefe T, Yue X. Seminars in cell & developmental biology. 2011;22:759. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 25.Boyer TG, Martin MED, Lees E, Riccardi RP, Berk AJ. Nature. 1999;399:276. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 26.Fondell JD, Ge H, Roeder RG. Proc. Natl. Acad. Sci. USA. 1996;93:8329. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hengartner CJ, Thompson CM, Zhang J, Chao DM, Liao S, Koleske AJ, Okamura S, Young RA. Genes Dev. 1995;9:897. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 28.Ito M, Yuan C, Malik S, Gu W, Fondell JD, Yamamura S, Fu Z, Zhang X, Qin J, Roeder RG. Mol. Cell. 1999;3:361. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y, Bjorklund S, Li Y, Sayre MH, Kornberg RD. Cell. 1994;77:599. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 30.Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Nature. 1999;398:828. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 31.Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Nature. 1999;398:824. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 32.Ryu S, Zhou S, Ladurner AG, Tjian R. Nature. 1999;397:446. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 33.Taatjes DJ, Naar AM, Andel F, Nogales E, Tjian R. Science. 2002;295:1058. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 34.Balamotis MA, Pennella MA, Stevens JL, Wasylyk B, Belmont AS, Berk AJ. Sci Signal. 2009;2:ra20. doi: 10.1126/scisignal.1164302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer KD, Lin S, Bernecky C, Gao Y, Taatjes DJ. Nat Struct Mol Biol. 2010;17:753. doi: 10.1038/nsmb.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hopkins AL, Groom CR. Nature reviews. Drug discovery. 2002;1:727. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 37.Fry DC. Current protein & peptide science. 2008;9:240. doi: 10.2174/138920308784533989. [DOI] [PubMed] [Google Scholar]
- 38.Zinzalla G, Thurston DE. Future medicinal chemistry. 2009;1:65. doi: 10.4155/fmc.09.12. [DOI] [PubMed] [Google Scholar]
- 39.Lariviere L, Seizl M, Cramer P. Curr Opin Cell Biol. 2012;24:305. doi: 10.1016/j.ceb.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun Z, Watts JL, DeBeaumont R, Mako Saito R, Hyberts SG, Yang S, Macol C, Iyer L, Tjian R, van den Heuvel S, Hart AC, Wagner G, Naar AM. Nature. 2006;442:700. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 41.Milbradt AG, Kulkarni M, Yi T, Takeuchi K, Sun ZY, Luna RE, Selenko P, Naar AM, Wagner G. Nat Struct Mol Biol. 2011;18:410. doi: 10.1038/nsmb.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vojnic E, Mourao A, Seizl M, Simon B, Wenzeck L, Lariviere L, Baumli S, Baumgart K, Meisterernst M, Sattler M, Cramer P. Nat Struct Mol Biol. 2011;18:404. doi: 10.1038/nsmb.1997. [DOI] [PubMed] [Google Scholar]
- 43.Taatjes DJ, Tjian R. Mol Cell. 2004;14:675. doi: 10.1016/j.molcel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Ito M, Okano HJ, Darnell RB, Roeder RG. EMBO J. 2002;21:3464. doi: 10.1093/emboj/cdf348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Nature. 2002;417:563. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 46.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Mol. Cell. 2000;5:683. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 47.Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument-Bromage H, Tempst P, Berger I, Kornberg GL, Asturias FJ, Kornberg RD, Takagi Y. Nature. 2011;475:240. doi: 10.1038/nature10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lariviere L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P. Nature. 2012;492:448. doi: 10.1038/nature11670. [DOI] [PubMed] [Google Scholar]
- 49.Robinson PJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. Proc Natl Acad Sci U S A. 2012;109:17931. doi: 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalkhoven E. Biochemical pharmacology. 2004;68:1145. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 51.Rowe SP, Mapp AK. Biopolymers. 2008;89:578. doi: 10.1002/bip.20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knuesel MT, Taatjes DJ. Transcription. 2011;2:28. doi: 10.4161/trns.2.1.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebmeier CC, Taatjes DJ. Proc Natl Acad Sci U S A. 2010;107:11283. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asada S, Choi Y, Uesugi M. J Am Chem Soc. 2003;125:4992. doi: 10.1021/ja0292703. [DOI] [PubMed] [Google Scholar]
- 55.Jung D, Shimogawa H, Kwon Y, Mao Q, Sato S, Kamisuki S, Kigoshi H, Uesugi M. J Am Chem Soc. 2009;131:4774. doi: 10.1021/ja900669k. [DOI] [PubMed] [Google Scholar]
- 56.Kwon Y, Arndt HD, Mao Q, Choi Y, Kawazoe Y, Dervan PB, Uesugi M. J Am Chem Soc. 2004;126:15940. doi: 10.1021/ja0445140. [DOI] [PubMed] [Google Scholar]
- 57.Wands AM, Wang N, Lum JK, Hsieh J, Fierke CA, Mapp AK. J Biol Chem. 2011;286:16238. doi: 10.1074/jbc.M110.207589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Z, Belanger G, Brennan BB, Lum JK, Minter AR, Rowe SP, Plachetka A, Majmudar CY, Mapp AK. J Am Chem Soc. 2003;125:12390. doi: 10.1021/ja036685v. [DOI] [PubMed] [Google Scholar]
- 59.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Nature. 2009;462:182. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. Science. 2004;303:844. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 61.Groner B. Peptides as Drugs. Mannheim: Wiley-VCH; 2009. [Google Scholar]
- 62.Benyamini H, Friedler A. Future medicinal chemistry. 2010;2:989. doi: 10.4155/fmc.10.196. [DOI] [PubMed] [Google Scholar]
- 63.Nielsen PE. Pseudo-peptides in Drug Discovery. Mannheim: Wiley-VCH; 2004. [Google Scholar]
- 64.Giniger E, Ptashne M. Nature. 1987;330:670. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 65.Buhrlage SJ, Bates CA, Rowe SP, Minter AR, Brennan BB, Majmudar CY, Wemmer DE, Al-Hashimi H, Mapp AK. ACS chemical biology. 2009;4:335. doi: 10.1021/cb900028j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buhrlage SJ, Brennan BB, Minter AR, Mapp AK. J Am Chem Soc. 2005;127:12456. doi: 10.1021/ja0536567. [DOI] [PubMed] [Google Scholar]
- 67.Minter AR, Brennan BB, Mapp AK. J Am Chem Soc. 2004;126:10504. doi: 10.1021/ja0473889. [DOI] [PubMed] [Google Scholar]
- 68.Andrews MJI, Tabor AB. Tetrahedron. 1999;55:11711. [Google Scholar]
- 69.Garner J, Harding MM. Organic & biomolecular chemistry. 2007;5:3577. doi: 10.1039/b710425a. [DOI] [PubMed] [Google Scholar]
- 70.Kritzer JA, Stephens OM, Guarracino DA, Reznik SK, Schepartz A. Bioorganic & medicinal chemistry. 2005;13:11. doi: 10.1016/j.bmc.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guarracino DA, Bullock BN, Arora PS. Biopolymers. 2011;95:1. doi: 10.1002/bip.21546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leduc AM, Trent JO, Wittliff JL, Bramlett KS, Briggs SL, Chirgadze NY, Wang Y, Burris TP, Spatola AF. Proc Natl Acad Sci U S A. 2003;100:11273. doi: 10.1073/pnas.1934759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Estieu-Gionnet K, Guichard G. Expert opinion on drug discovery. 2011;6:937. doi: 10.1517/17460441.2011.603723. [DOI] [PubMed] [Google Scholar]
- 74.Schafmeister CE, Po J, Verdine GL. J Am Chem Soc. 2000;122:5891. [Google Scholar]
- 75.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. J Am Chem Soc. 2007;129:2456. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phillips C, Roberts LR, Schade M, Bazin R, Bent A, Davies NL, Moore R, Pannifer AD, Pickford AR, Prior SH, Read CM, Scott A, Brown DG, Xu B, Irving SL. J Am Chem Soc. 2011;133:9696. doi: 10.1021/ja202946k. [DOI] [PubMed] [Google Scholar]
- 78.Baek S, Kutchukian PS, Verdine GL, Huber R, Holak TA, Lee KW, Popowicz GM. J Am Chem Soc. 2012;134:103. doi: 10.1021/ja2090367. [DOI] [PubMed] [Google Scholar]
- 79.Wells JA, McClendon CL. Nature. 2007;450:1001. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 80.Schreiber SL. Science. 2000;287:1964. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 81.Spandl RJ, Bender A, Spring DR. Organic & biomolecular chemistry. 2008;6:1149. doi: 10.1039/b719372f. [DOI] [PubMed] [Google Scholar]
- 82.Koehler AN, Shamji AF, Schreiber SL. J Am Chem Soc. 2003;125:8420. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]
- 83.Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. Nature. 2002;416:653. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- 84.Ng PY, Tang Y, Knosp WM, Stadler HS, Shaw JT. Angew Chem Int Ed Engl. 2007;46:5352. doi: 10.1002/anie.200700762. [DOI] [PubMed] [Google Scholar]
- 85.Scott DE, Coyne AG, Hudson SA, Abell C. Biochemistry. 2012;51:4990. doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- 86.Scott DE, Ehebauer MT, Pukala T, Marsh M, Blundell TL, Venkitaraman AR, Abell C, Hyvonen M. Chembiochem : a European journal of chemical biology. 2013;14:332. doi: 10.1002/cbic.201200521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bruncko M, Oost TK, Belli BA, Ding H, Joseph MK, Kunzer A, Martineau D, McClellan WJ, Mitten M, Ng SC, Nimmer PM, Oltersdorf T, Park CM, Petros AM, Shoemaker AR, Song X, Wang X, Wendt MD, Zhang H, Fesik SW, Rosenberg SH, Elmore SW. J Med Chem. 2007;50:641. doi: 10.1021/jm061152t. [DOI] [PubMed] [Google Scholar]
- 88.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, Roberts L, Tahir SK, Xiao Y, Yang X, Zhang H, Fesik S, Rosenberg SH, Elmore SW. Cancer Res. 2008;68:3421. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 89.Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit RE, Hahn S. Mol Cell. 2011;44:942. doi: 10.1016/j.molcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Herbig E, Warfield L, Fish L, Fishburn J, Knutson BA, Moorefield B, Pacheco D, Hahn S. Mol Cell Biol. 2010;30:2376. doi: 10.1128/MCB.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jedidi I, Zhang F, Qiu H, Stahl SJ, Palmer I, Kaufman JD, Nadaud PS, Mukherjee S, Wingfield PT, Jaroniec CP, Hinnebusch AG. J Biol Chem. 2010;285:2438. doi: 10.1074/jbc.M109.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]