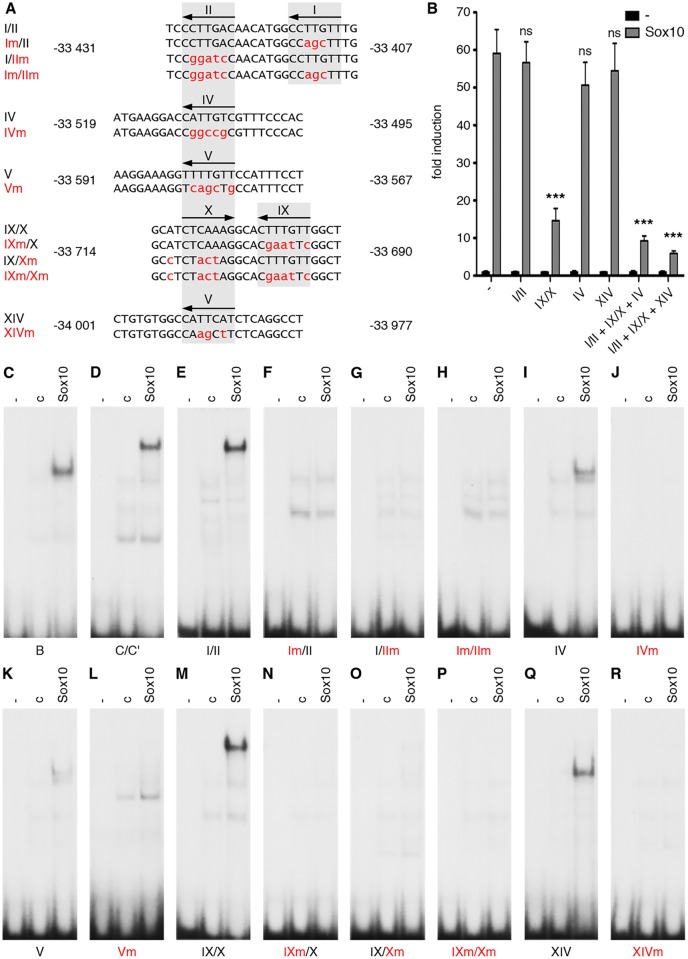
Fig 8. Sox binding sites mediate OLEa responsiveness to transcriptional activation by Sox10.
(A) Three or more nucleotides (small red letters) were changed within the Sox binding sites I/II, IV, V, IX/X and XIV of OLEa to generate mutant sites. Numbers on both sides of the sequence indicate the exact position relative to the transcriptional start. Location of Sox sites are highlighted in grey and orientations indicated by arrows. (B) Sox binding site mutations were introduced in several combinations into the OLEa reporter plasmid and luciferase assays were performed after transient transfection of this plasmid in Neuro2A cells in the absence or presence of Sox10 expression plasmid. The Sox binding sites mutated in each reporter construct are given below the lanes. Luciferase activities were determined in four experiments each performed in duplicates. The activity obtained for each luciferase reporter in the absence of Sox10 was arbitrarily set to 1. Fold inductions for Sox10 were calculated in relation and are presented as mean ± SEM. Statistical significance of fold inductions obtained with mutant reporters relative to wildtype was determined by one way analysis of variance (ANOVA) with Tukey’s multiple comparison post test (ns, not significant; ***, P ≤0.001). (C-R) EMSA was performed with radiolabelled double-stranded oligonucleotides containing Sox binding sites in wildtype (same gels as in Fig. 6) or mutant version. Oligonucleotides were incubated in the absence (-), or presence (c, Sox10) of protein extracts before gel electrophoresis as indicated above the lanes. Extracts were from mock-transfected HEK293 cells (c) or HEK293 cells expressing full length Sox10 (Sox10). Oligonucleotides with site B and site C/C‘ from the promoter of the Mpz (myelin protein zero) gene served as positive control for Sox10 monomer and dimer binding.