Abstract
In cyanobacterial Nostoc species, substratum-dependent gliding motility is confined to specialized nongrowing filaments called hormogonia, which differentiate from vegetative filaments as part of a conditional life cycle and function as dispersal units. Here we confirm that Nostoc punctiforme hormogonia are positively phototactic to white light over a wide range of intensities. N. punctiforme contains two gene clusters (clusters 2 and 2i), each of which encodes modular cyanobacteriochrome–methyl-accepting chemotaxis proteins (MCPs) and other proteins that putatively constitute a basic chemotaxis-like signal transduction complex. Transcriptional analysis established that all genes in clusters 2 and 2i, plus two additional clusters (clusters 1 and 3) with genes encoding MCPs lacking cyanobacteriochrome sensory domains, are upregulated during the differentiation of hormogonia. Mutational analysis determined that only genes in cluster 2i are essential for positive phototaxis in N. punctiforme hormogonia; here these genes are designated ptx (for phototaxis) genes. The cluster is unusual in containing complete or partial duplicates of genes encoding proteins homologous to the well-described chemotaxis elements CheY, CheW, MCP, and CheA. The cyanobacteriochrome-MCP gene (ptxD) lacks transmembrane domains and has 7 potential binding sites for bilins. The transcriptional start site of the ptx genes does not resemble a sigma 70 consensus recognition sequence; moreover, it is upstream of two genes encoding gas vesicle proteins (gvpA and gvpC), which also are expressed only in the hormogonium filaments of N. punctiforme.
INTRODUCTION
Nearly half of unicellular and more than three-quarters of filamentous cyanobacteria in culture are motile by a gliding mechanism on a solid substratum (1). Gliding motility is a property that is rapidly lost during prolonged serial transfer in culture (2, 3, 4). In members of the nitrogen-fixing, heterocyst-forming cyanobacterial taxonomic subsection IV (Nostocales), gliding motility, if present, occurs either by vegetative filaments (in the genera Anabaena, Aphanizomenon, Cylindrospermum, and others) or exclusively by specifically differentiated filaments called hormogonia (in the genera Calothrix, Nostoc, Tolypothrix, and others) (5). Substratum-dependent gliding is much slower than flagellar swimming in suspension, ranging from 5 μm/min in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 (6) to 18 μm/min by vegetative filaments of the heterocyst-forming cyanobacterium Anabaena cylindrica (7), 60 μm/min by hormogonium filaments of Nostoc cycadae (8), and 600 μm/min by the filamentous non-heterocyst-forming cyanobacterium Oscillatoria princeps (9).
Phototaxis is a behavioral characteristic of Bacteria, Archaea, and single-cell eukaryotes, as well as some animal larvae (10) that contain photoreceptors. It is a positive selective property for a phototrophic organism, allowing movement both toward higher light intensities for maximal photosynthetic activity and away from inhibitory high intensities or wavelengths that could result in photooxidative stress or other damage. Directional gliding motility in cyanobacteria has long been observed, but only recently, through genetic analysis, have the mechanisms of motility and photoperception been identified in the unicellular organism Synechocystis sp. strain PCC 6803 (for a review, see reference 11). Gliding by Synechocystis PCC 6803 is mediated by the extrusion and retraction of type IV pili under the control of PilB and PilT proteins, respectively (11). Pilin synthesis is under the control of a chemotaxis-like signal transduction system called either Tax3 (12) or Pil (13). The genome organization in Synechocystis PCC 6803 is unusual in that the gene encoding the homolog of the histidine protein kinase CheA is separate from the other chemotaxis protein-encoding genes and is fragmented into two genes, one encoding the C-terminal ATP binding and catalytic domain (slr0322) and the other encoding the N-terminal histidine phosphotransfer (Hpt) domain (slr0073) (13).
A photoreceptor (encoded by sll0041) that is required for positive phototaxis by Synechocystis PCC 6803 is a cyanobacteriochrome. Cyanobacteriochromes are bilin binding proteins with homology to the chromophore-binding domains of phytochromes and are associated with various output domains, such as histidine kinase domains and methyl-accepting chemotaxis protein (MCP) signaling domains (14). In Synechocystis PCC 6803, the sensory cyanobacteriochrome domain is encoded by a gene (sll0041) that is separate from the gene encoding the MCP signaling domain (sll0042). However, both genes are in a cluster of genes encoding proteins homologous to the well-described CheY, CheW, and CheA proteins of a chemotaxis signal transduction complex (12, 13). The cluster and individual genes in Synechocystis PCC 6803 have been differently named by different investigators (12, 13, 15); based on the body of published reports, we follow the pix designation for the cluster and genes (13).
The name pixJ has been assigned to a gene (all1069) in Nostoc (Anabaena) sp. strain PCC 7120 encoding the complete cyanobacteriochrome-MCP homologous to the Synechocystis PCC 6803 PixJ1 (sll0041) and PixJ2 (sll0042) proteins (16). It is also localized in a cluster of genes encoding chemotaxis-like proteins. Since Nostoc PCC 7120 no longer differentiates hormogonia and is nonmotile, functional assays of PixJ in the context of phototaxis in this organism are not possible, although biochemical studies of bilin binding domains of the protein expressed in Escherichia coli have been reported (16).
We initiated studies on motility and taxis by hormogonium filaments of the N2-fixing, symbiotically competent Nostoc punctiforme strain ATCC 29133. Hormogonia are nongrowing dispersal units whose cells are often smaller and differently shaped than parental filament vegetative cells (1). Hormogonia differentiate transiently in response to a variety of positive or negative factors for growth, including, in symbiotically competent strains, chemical factors produced by symbiotic plant partners (17). The formation of hormogonia in N. punctiforme involves differential transcription of as many as 2,900 genes (ca. 43% of the protein-encoding genome) over a 24-h period (18); thus, hormogonia are a dynamic part of the life and developmental cycle of N. punctiforme. To our knowledge, there have been no studies confirming phototactic behavior in the hormogonia of any cyanobacterium.
Two gene clusters have been identified as essential for N. punctiforme hormogonium motility (19, 20). One cluster encodes chemotaxis-like proteins (20), among which the CheA protein is phylogenetically homologous to Synechocystis PCC 6803 CheA (encoded by slr0322 and slr0073) (21) and is involved in the regulation of the expression of genes encoding the pilin motility motor (13). However, in contrast to the organization of the cluster in Synechocystis PCC 6803, all of these genes are in an operon in N. punctiforme, and the CheA-like histidine kinase is encoded by a single gene (20). Deletion of the entire cluster in N. punctiforme results in a nonmotile phenotype, where the hormogonium cells fail to secrete extracellular polysaccharide. The gene cluster is designated the hmp locus (for hormogonium motility and polysaccharide) (20) (see Table 1).
TABLE 1.
N. punctiforme taxis-like gene loci
| Cluster | Locus taga | Gene designation | Homologyb |
|---|---|---|---|
| 1 | NpF5960 | hmpA | CheY, PatA family |
| NpF5961 | hmpB | CheY | |
| NpF5962 | hmpC | CheW | |
| NpF5963 | hmpD | MCP | |
| NpF5964 | hmpE | CheA | |
| 2 | NpR6010 | pixL | CheA |
| NpR6012 | pixJ | MCP | |
| NpR6013 | pixI | CheW | |
| NpR6014 | pixH | CheY | |
| NpR6015 | pixG | CheY, PatA family | |
| 2i | NpF2161 | ptxA | CheY, PatA family |
| NpF2162 | ptxB | CheY | |
| NpF2163 | ptxC | CheW | |
| NpF2164 | ptxD | MCP | |
| NpF2165 | ptxE | CheA | |
| NpF2166 | ptxF | CheW | |
| NpF2167 | ptxG | MCP fragment | |
| NpF2168 | ptxH | CheA fragment | |
| 3 | NpF5636 | CheY, PatA family | |
| NpF5637 | CheY | ||
| NpF5638 | CheW | ||
| NpF5639 | MCP | ||
| NpF5640 | CheA | ||
| L | NpR0244 | CheB | |
| NpR0245 | CheA | ||
| NpR0246 | CheW | ||
| NpR0247 | MCP | ||
| NpR0248 | CheR | ||
| NpR0249 | MCP | ||
| NpR0250 | CheW |
The locus tags presented here are from the original database constructed for the N. punctiforme genome and used for system-level analyses. Examples of renamed locus tags in the NCBI database are Npun_F0001 and Npun_R0003.
Homology to taxis proteins identified in multiple heterotrophic bacteria.
Comparative transcriptomics and further mutational analysis established that the Hmp signal transduction system regulates the transcription of a second cluster of genes, designated hps (for hormogonium polysaccharide) genes, because the gene products are involved in the synthesis of a polysaccharide(s) and the basic components of a polysaccharide secretion system (19). The core proteins of the secretion system are computationally annotated as pseudopilins and conserved hypothetical proteins; the system most likely represents the junctional pores seen in electron micrographs and suggested to constitute the nozzles for polysaccharide secretion required for gliding (22). Although polysaccharide secretion is essential for hormogonium gliding motility, mutation analysis also implicates type IV pili (23). Therefore, the actual motor in filamentous gliding has yet to be unequivocally identified. There is similarity in the functions of two homologous chemotaxis-like signal transduction complexes in unicellular and filamentous cyanobacteria in regulating the expression of different motility motors.
In addition to the hmp locus, the genome of N. punctiforme contains four other loci of genes encoding chemotaxis-like proteins (Table 1). One locus (NpR0244 to NpR0250) appears to have originated by horizontal gene transfer, while the hmp locus and the other three loci evolved in the cyanobacterial lineage (21). We report here that N. punctiforme hormogonia display a phototactic response to white light, and we identify the gene locus responsible for the response.
MATERIALS AND METHODS
Cultures and culture conditions.
All experiments reported in this paper were conducted with wild-type N. punctiforme strain ATCC 29133 (UCD 154), which displays the morphology and physiology of the original isolate (strain PCC 73102), and mutants derived from it. Wild-type and mutant N. punctiforme strains were grown under standard culture conditions as described previously (24) in liquid Allen and Arnon (AA) medium (25) diluted 4-fold (AA/4) and on solidified AA medium plates. When necessary, the medium was supplemented with 2.5 mM NH4Cl buffered with 5 mM morpholinepropanesulfonic acid (MOPS; pH 7.8). Neomycin at 25 μg/ml, chloramphenicol at 30 μg/ml, and ampicillin at 10 μg/ml were used for the selection and maintenance of recombinant N. punctiforme strains. Escherichia coli strains and derivatives were grown in lysogeny broth (LB) supplemented with antibiotics at the following concentrations: kanamycin, 25 μg/ml; chloramphenicol, 30 μg/ml; ampicillin, 100 μg/ml.
The symbiotic plant partner hornwort (Anthoceros punctatus L.) was cultured under standard conditions (26) in Hutner's medium containing ammonium nitrate as a nitrogen source and supplemented with 0.5% (wt/vol) glucose. A plant exudate containing a hormogonium-inducing factor (HIF) was obtained and harvested according to the method in reference 18, except that A. punctatus cultures were starved of nitrogen in plant growth medium for a shorter time, 4 to 7 days. N. punctiforme filaments were suspended in the HIF solution after the removal of plant tissue and were incubated overnight under standard conditions for hormogonium induction. Upon the production of motile N. punctiforme hormogonia, verified microscopically, the cultures were immediately used for phototaxis assays or were harvested by centrifugation at 14,000 × g for 5 min, quick-frozen in liquid nitrogen, and then stored at −80°C for subsequent RNA extraction.
Phototaxis assays.
Square petri plates (Fisher) containing growth medium solidified with 0.5% (wt/vol) Noble agar were covered with a quartz glass plate to allow unimpeded penetration of light of all wavelengths and to prevent the agar plate from drying out during incubation. The whole petri plate except for 1.5 cm at one end was obscured with black tape to prevent light penetration. The edges of the petri plates were also taped with black tape, so light was allowed to reach the plate only at the 1.5-cm untaped area at one edge, allowing the application of directional lighting from the top face at one edge of the plate. Motile N. punctiforme hormogonium filaments obtained by overnight induction with HIF were collected by brief centrifugation (16,000 × g for 30 s) and were placed 4 cm back from the lighted edge of the petri plate (approximately the middle of the plate as indicated by grid marks on the bottom of the plate). As much liquid medium was removed as possible, so the cells were directly in contact with the agar surface. Efforts were made to use similar amounts of cells in each experiment; however, since the induced cultures clump, and clump sizes differ, accurate biomass measurements were not routinely possible. Moreover, the hormogonium filaments adhered to plastic pipette tips, such that it was not always possible to dislodge all of the filaments. Thus prepared, the covered plates were incubated, in a “black box” with a light source pointed at the unobscured end of the petri plate, at room temperature for 24 h to allow taxis to occur (see Fig. S1 in the supplemental material). Light intensities were determined by using a light meter (S130C; Thorlabs) placed on top of the untaped portion of the quartz glass plate. For “white-light” LED (light-emitting diode) experiments, the LED (LEDH-5500; Lightspeed Technologies) was covered with a yellow 101 filter (Lee Filters), which blocks emissions below ∼475 nm and effectively negates the 455-nm pump peak that is characteristic of white-light LEDs. Under these conditions, LEDs did not significantly increase the temperature in the black box. For high-intensity white-light experiments, a tungsten light source from a 45-W, 120-V, 330-lm flood lamp was used; fans were provided to help cool the incubation chamber, and in all the experiments reported here, temperatures did not exceed 30°C. A Nikon D50 camera captured images of each plate at 0 and 24 h after incubation in the light box. ImageJ software was used to quantify the integrated density (the product of the area and the mean gray value) of each phototaxis experiment in order to determine the amount of movement of filaments toward or away from the light (see Fig. S2 in the supplemental material). Multiple biological replicates were performed for each of the experiments reported here; however, due to biomass variability (see above), only a representative picture of the results of each of the experimental conditions is shown and quantified. All of the photos presented here are oriented such that the light was applied from the bottom of the photograph.
Transcription analysis.
Analysis of DNA microarray data from N. punctiforme strain 29133S (UCD 153) HIF-induced hormogonia (18) showed that the level of transcription of locus 2i (N. punctiforme chemotaxis-like loci numbered according to reference 21) increased between 0.5 and 24 h after induction. Thus, RNA was isolated from frozen samples of N. punctiforme ATCC 29133 (UCD 154) collected at 1, 3, 6, 12, 18, and 24 h after HIF induction. The time course of expression of genes in the 5 chemotaxis-like loci was determined by analyses of the DNA microarray data from reference 19, which were conducted as described previously (24). The original array data from reference 19 are deposited in the Gene Expression Omnibus database under accession no. GSE42859. The data were normalized and analyzed in the R statistical platform and were collected and plotted in Excel; they are summarized here (see Table S1 in the supplemental material).
The RNA samples collected at 12 h after HIF induction were used in the subsequent 5′ RACE (rapid amplification of 5′ cDNA ends) assays that allowed the identification of the N. punctiforme transcriptional start site (TSS) for locus 2i. The experiments were carried out initially for all eight genes of locus 2i, plus the two upstream gvp genes, using 1 μg of total RNA and the 5′ RACE System for Rapid Amplification of cDNA Ends, version 2.0, according to the manual from Invitrogen, except that the first-strand cDNA synthesis was performed using SuperScript III reverse transcriptase (Invitrogen), and the poly(dC) of cDNA was synthesized using a terminal deoxynucleotidyl transferase (TdT) from New England BioLabs (according to the manufacturer's instructions). Since the products from all 10 reactions were consistent with 1 TSS and 1 downstream 5′ end, only the ptxA and ptxH reactions are summarized here. In order to discern the difference between a TSS and a processed 5′ end, a modified 5′ RACE protocol using tobacco acid pyrophosphatase (TAP)-treated RNA (according to references 27 and 28) was performed. TAP hydrolyzes the 5′ triphosphate of unprocessed transcripts, allowing the ligation of an adaptor. Comparison of the products from PCR amplifications from a TAP-treated and a non-TAP-treated RNA allows the differentiation of unprocessed, legitimate TSS from processed 5′ ends. For these assays, 20 μg of total RNA was used with the enzymes TAP (Epicentre Technologies), T4 RNA ligase (New England BioLabs), and SuperScript III reverse transcriptase (Invitrogen), according to the respective manufacturers' instructions. The gene-specific primers were used together with the kit adaptor-specific primers in PCR amplifications carried out with the following profiles: 94°C for 5 min, followed by 35 cycles of 30 s at 95°C, 30 s at 55°C or 58°C, and 1 min at 72°C, concluding with a 7-min extension at 72°C. The PCR products obtained were sequenced at the UC Davis DNA Sequencing Facility. The ptxA reactions were performed using primers NpF2161-gsp1 (5′-CTGCATGATATCAAACAGGACTTCAG-3′) and NpF2161-gsp2 (5′-GCTATTGAGCTTGCAATCAGATAAC-3′). The ptxH reactions were performed using primers NpF2168-gsp1 (5′-CCGATACTGATGCAGAAGCTG-3′) and NpF2168-gsp2 (5′-CAACTATAGCAATGGCATC-3′).
Construction of mutants.
All chromosomal mutations were introduced into wild-type N. punctiforme via triparental conjugation using E. coli strains as carriers of recombinant plasmids according to reference 29. Mutant N. punctiforme strains were selected by the presence of neomycin, and single recombinant strains, in which the suicide plasmid was integrated into the genome, were maintained in a medium containing neomycin and chloramphenicol, or neomycin only, for sacB-containing pRL271- and pRL278-derived plasmids (30), respectively. Strains in which the suicide plasmid and either the wild-type or the mutant gene were eliminated via a second recombination event were selected by the presence of 5% (wt/vol) sucrose. PCR and/or restriction mapping followed by Southern hybridization confirmed the elimination of the wild-type gene and its replacement with the inactivated gene (data not shown). All insertion mutants were constructed by first isolating a large fragment of genomic DNA from a sequencing fosmid that contained the gene of interest along with sufficient adjacent DNA (≥1.0 kb) to allow for homologous recombination. The antibiotic cassette, Ω-npt, from pSCR9 (31) was then inserted into the gene of interest to interrupt the reading frame, except for strain UCD 520, in which a portion (862 bp) of the gene was deleted at the same time as the insertion, creating a deletion-insertion. Because the Ω-npt fragment contains transcriptional and translational stops in all three reading frames, insertional mutations have negative polar effects on downstream genes. All in-frame deletion mutants were constructed by PCR. Primers were designed to amplify DNA to the right and left of the deletion, with the primers adjacent to the deletion containing overlapping sequences. The PCR products were mixed and were allowed to anneal via overlapping sequences, and a subsequent PCR produced a product with the deletion. All PCR products were sequenced to verify that the only mutation was the deletion. These mutations should affect only the deleted portion and should not have a polar effect on downstream genes. The details for each construct are given in Table 2.
TABLE 2.
Details of the construction of N. punctiforme mutants
| Mutant N. punctiforme strain | Mutation (locus tag) | Construction details |
|---|---|---|
| UCD 510 | NpF5639::Ω-npt (NpF5639) | Ω-npt was inserted into the Eco47III site on a 5.17-kb XbaI fragment from fosmid fla2304 |
| UCD 512 | ptxD::Ω-npt (NpF2164) | Ω-npt was inserted into the NheI site on a 7.2-kb ScaI fragment from fosmid fla0741 |
| UCD 520 | ptxHΔ::Ω-npt (NpF2168) | Ω-npt replaced an 862-bp EcoRI fragment on a 7.8-kb NruI-HpaI fragment from fosmid fla0741 |
| UCD 543 | hmpDΔ (NpF5963) | Previously published (21); mutation creates a deletion of 2,934 bp |
| UCD 549 | gvpA::Ω-npt (NpF2159) | Ω-npt was inserted into the AflIII site on a 4.2-kb ScaI fragment from fosmid fla0741 |
| UCD 598 | ptxEΔ (NpF2165) | Primers cheAL (CGATCGCTCCGATCTTTATTGG), cheAI1 (GTCGCTCAATTGGTGCAACACAG), cheAI3 (CCAAGCGTTATACGCTCCAGAAGTCGCTCAATTGGTGCAACACAG), cheAR (GTGCAGCCGCAACTATAGCAATG) |
| UCD 571 | cheLΔ (NpR0244–0250) | Primers NpR0250-5′-BglII-F (ATATAAGATCTATATGCAAGACAACTCAAAAATTGATC), NpR0250-5′-SmaI-R (GGTTATCGATTCCCGGGCGATTTGGATACCAATCATTC), NpR0244-3′-SmaI-F (GTATCCAAATCGCCCGGGAATCGATAACCAATAACAACTTTGC), NpR0244-3′-SacI-R (ATATAGAGCTCATTGCTCAGACTTGGAAAGCTG); mutation creates a deletion of 9,830 bp |
| UCD 623 | pixJΔ (NpR6012) | Primers P442L (CGGTAGTAACAGGAGGATAG), P443R (GTTAATCAAATGACATTTTTGTTTAAAGTCAAATAGATTTTAGATTTTGG), P444R (CAAAAATGTCATTTGATTAAC), P445L (AAATTCTGTCCGCTACGTAC); mutation creates a deletion of 3,282 bp |
RESULTS
Phototactic response.
N. punctiforme strain ATCC 29133 (UCD 154) has the morphology and life cycle characteristics of the organism originally isolated: it differentiates a basal level (ca. 10% of the filaments) of hormogonia during the exponential and light-limited linear growth phases with combined nitrogen or N2 as the nitrogen source; the hormogonia stimulate clumping with vegetative filaments, such that the culture grows as colonial aggregates of different sizes (20); and the hormogonia show positive taxis to white light (Fig. 1). These characteristics contrast with those of a spontaneous mutant (strain UCD 153), which no longer differentiates hormogonia during exponential and linear growth but does so under inducing conditions. Strain UCD 153 shows dispersed growth under shaking culture conditions and, while motile and symbiotically competent, does not exhibit a phototactic response in plate assays (data not shown). Strain UCD 153 was used in experiments to define the transcriptome during hormogonium differentiation, because prior to induction, the culture contains no hormogonia (18). The frequency of gene replacement by conjugal transfer is 1 or 2 orders of magnitude lower in strain UCD 154 than in strain UCD 153. However, unlike strain UCD 153, strain UCD 154 exhibits phototaxis and was thus used for genetic analysis of this process.
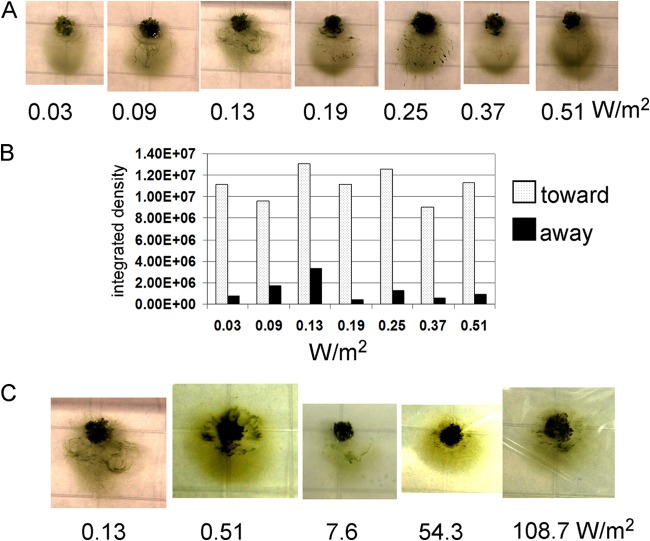
FIG 1.
(A) Images of hormogonium filaments of wild-type N. punctiforme (strain UCD 154) migrating on representative phototaxis plates after 24 h of directed-light incubation using white-light LEDs. The light was applied from the direction of the bottom of the photograph. The intensity of the light used is given below each panel. (B) Quantification (see supplemental information) of each of the images in panel A. Cells moving downward in the images are scored as moving toward light, and cells moving upward in the images are scored as moving away from light. Integrated density values were derived from the ImageJ analysis software. (C) Images of phototaxis plates incubated under tungsten lamps, except for the first picture on the left, of a plate incubated with a white LED light, which is included here as a control for purposes of comparison. The intensity of the light applied is given below each image. The direction of light was from the bottom of the photograph.
Hormogonium filaments glide, individually or as a mass of filaments, as seen in microscopic observations (19) (data not shown); there is no apparent requirement for filament-to-filament contact. On an agar surface, the filaments show phototactic movement from the point of inoculation toward the light source in a fan-like pattern. Masses of filaments may turn sideways to the light source such that a ripple pattern forms (Fig. 1A). The phototactic response of N. punctiforme hormogonia has a distinctly low light threshold for activation, less than 0.03 W/m2 (0.13 μmol photons/m2/s) (Fig. 1A), and the response does not markedly increase at light increments up to a 4.5-fold-higher intensity, as determined by quantification of the movement of filaments toward or away from the point of inoculation (Fig. 1B). Since our LED light system reached maximal intensity at 0.5 W/m2, we switched to a 45-W tungsten light source to achieve higher light intensities, with cooling fans to minimize heat buildup. The filaments displayed positive phototaxis under these light conditions at intensities from 0.51 W/m2 (2.35 μmol/m2/s) to 108.7 W/m2(500 μmol/m2/s) (Fig. 1C), with no obvious light intensity-dependent increase or decrease in distance of movement. At 108.7 W/m2, there was substantial drying of the agar plate due to the increased air circulation required to maintain a temperature below 30°C, but negative phototaxis, or net movement away from light, was never observed in these high-light experiments.
Signal transduction systems.
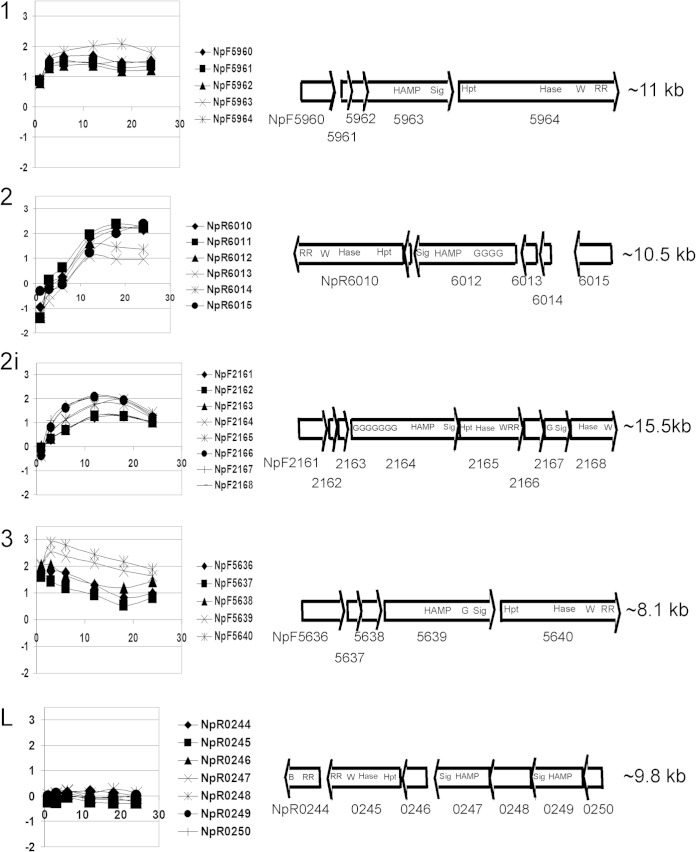
Since the light receptor for phototaxis in Synechocystis PCC 6803 is a cyanobacteriochrome, we searched the N. punctiforme genome for genes encoding homologous proteins. N. punctiforme encodes 2 knotted and 4 knotless phytochromes and 16 cyanobacteriochromes (32). Two of the cyanobacteriochromes are associated with MCP signaling domains in two of the five clusters of genes encoding chemotaxis-like proteins (Fig. 2; summarized in Table 1). These clusters encoding chemotaxis-like proteins were previously recognized and computationally analyzed by Wuichet and Zhulin (21); we adopt their numerical identification system for loci. Four of the loci, loci 1, 2, 2i, and 3 (Fig. 2), evolved in the cyanobacterial lineage, while locus L was acquired by horizontal gene transfer (21). The genes encoding cyanobacteriochrome-MCPs are in loci 2 and 2i, which, based on information presented below, we have designated pix (with homology to pixJ in Synechocystis PCC 6803 and Nostoc PCC 7120) and ptx (for phototaxis), respectively (Table 1). Computational analyses imply that the cyanobacteriochrome-MCPs in loci 2 (PixJ) and 2i (PtxD) lack transmembrane domains and appear to be soluble in the cytoplasm. The MCP sensing domain in cluster 3 contains a GAF (cyclic GMP [cGMP]-specific phosphodiesterase, adenylyl cyclase, FhlA) domain and is computationally annotated as a phytochrome; however, this GAF domain lacks the two cysteines that bind bilin and is unlikely to participate in photoreception. The latter conclusion is based on experiments with the GAF 1 domain of the N. punctiforme cyanobacteriochrome encoded by NpF1883, which also lacks the conserved cysteines and failed to bind bilin in vivo and in vitro (33).
FIG 2.
The 5 chemotaxis-like loci. Numerical designations for each locus (to the left of each plot) are based on the system of Wuichet and Zhulin (21). (Left) Graphs of microarray data. The x axis values are hours after HIF induction, and the y axis values are expression levels (log base 2) of genes at each time point relative to expression at time zero. (Right) Open reading frame maps. The relative sizes of genes in each cluster are drawn to scale, but not all maps reflect the same size relative to each other. The approximate size of each locus is given on the right. The following abbreviations are used for the domains: HAMP, histidine kinase, adenylate cyclase, methyl-accepting protein, phosphatase; Sig, signaling domain; Hpt, histidine-containing phosphotransfer domain; Hase, histidine kinase; W, CheW binding domain; RR, response regulator receiver domain; G, GAF (cGMP-specific phosphodiesterase, adenylyl cyclase, FhlA); B, CheB methylesterase domain.
Motility occurs only in hormogonia of N. punctiforme; therefore, if gene products from any of these loci are involved in directed motility, the genes should be transcribed in hormogonia. The temporal transcription patterns following hormogonium induction by HIF in all five clusters are shown in Fig. 2. All of the genes in the four clusters of the cyanobacterial lineage were upregulated immediately following induction. Genes encoding CheY and CheW proteins (Table 1) in each of the four cyanobacterial-lineage clusters showed transcription signals slightly above background in the vegetative filament samples at time zero (data not shown). Thus, differential transcription cannot specifically identify which, if any, chemotaxis-like cluster might encode functional phototaxis proteins. However, the HIF induction followed two kinetic patterns. The genes in clusters 1 and 3 were immediately upregulated to the maximum extent within 3 h after induction. In contrast, transcription of the genes in clusters 2 and 2i increased at a lower rate, reaching peak expression at around 12 to 18 h. These patterns may have implications for the roles of the gene products in hormogonium formation and function. The gene products from locus 1 are now known to be required for the synthesis of a polysaccharide secretion system essential for motility (20), and the transcripts appear to accumulate well before those of loci 2 and 2i. Microarray data from a PCR-based internal gene fragment platform using strain UCD 153 indicated that the NpR0244 and NpR0247 genes in locus L, encoding CheB and MCP homologs, respectively, were upregulated less than 2-fold by 18 h following hormogonium induction (18). In the NimbleGen oligonucleotide-based array used here, none of the genes in locus L appear upregulated following induction by HIF.
To definitively identify genes and gene clusters involved in directed motility, we initiated a mutant analysis, with an initial focus primarily on genes encoding MCPs. The clusters appear to be organized and transcribed as operons. Thus, insertion mutations with the Ω-npt cassette most likely have a polar effect on the expression of downstream genes; therefore, where possible, we constructed in-frame deletions. Mutants with individual in-frame deletions of each of the genes, except for one, in the hmp cluster (locus 1), or with an in-frame deletion of the entire locus, differentiate nonmotile hormogonia (20) that do not respond by movement to light. The exception is a mutant with a deletion of NpF5960, encoding the PatA family CheY homolog (HmpA), which is motile and phototactic. The mutant (strain UCD 510) with an insertion in NpF5639, encoding the MCP-like protein in locus 3, differentiated hormogonium filaments that have a curved “S” shape rather than a straight morphology and a greatly reduced rate of gliding. Vegetative filaments of this mutant line have normal cell morphology and growth properties (data not shown). The abnormally shaped hormogonia, however, continue to move toward light (Fig. 3), although the distance they travel is much less than that for the wild type. We were unable to mutate the putatively marginally expressed MCP (encoded by NpR0247) in cluster L by gene replacement; therefore, we constructed a deletion of the entire cluster from NpR0244 to NpR0250. This mutant (strain UCD 571) differentiated hormogonia with wild-type morphology and rates of phototactic gliding motility similar to those of the wild type (Fig. 3). Hormogonia of the MCP (PixJ, encoded by NpR6012) in-frame deletion mutant in cluster 2 (strain UCD 623) also showed normal white-light phototaxis behavior. Only the cluster 2i MCP (PtxD, encoded by NpF2164) insertion mutant (strain UCD 512) failed to display directed net movement toward light (Fig. 3). Mutant strain UCD 512 was highly motile and showed approximately equal amounts of movement toward light as away from light (Fig. 3). Based on these results, we have named this gene cluster ptx (for phototaxis). If other photoreceptors are involved in directed motility, it appears that they must feed their information into the Ptx signal transduction system. All of the chemotaxis-like gene mutants analyzed, except for those in the hmp cluster, which are nonmotile, are symbiotically competent. They infected the hornwort A. punctatus under shaking liquid culture conditions at a frequency similar to that of the wild type and supported N2-dependent plant growth (data not shown).
FIG 3.
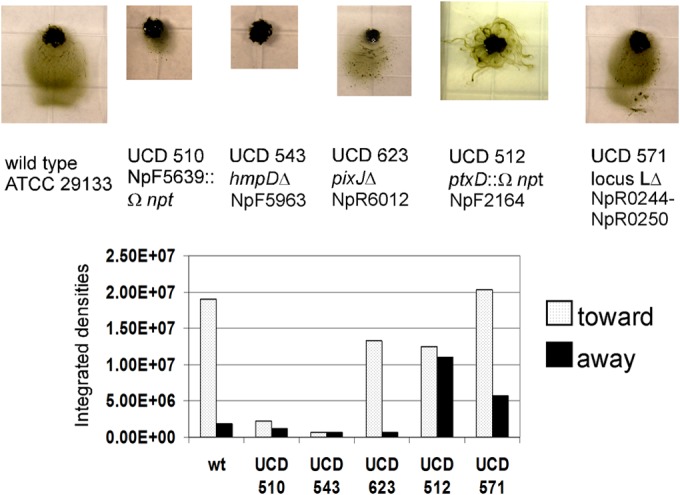
(Top) Representative phototaxis images of the wild-type strain (wt) and mutant strains with alterations in each of the 5 chemotaxis-like loci, incubated with white-light LEDs at 0.130 W/m2 for 24 h. (Bottom) Quantification of each of the images.
Transcription of genes in the ptx locus and requirement for gene products.
The ptx locus consists of eight genes encoding chemotaxis-like proteins (Table 1; Fig. 4). We employed 5′ RACE to identify the 5′ ends of the mRNAs and TAP-modified 5′ RACE to locate the transcriptional start site(s) in the locus. TAP removes the triphosphate from a 5′ end generated by RNA polymerase, such that the adaptor can be ligated. TAP had no effect on the ligation of the adaptor to the ends of cDNAs generated by primers to ptxH (Fig. 4), suggesting that these are processed ends. These results indicate the presence of an mRNA-processing site immediately 5′ of ptxGH. However, dephosphorylation did allow ligation to the end generated by ptxA primers, indicating a TSS (Fig. 4). Sequencing of the ptxA extended product revealed that the 5′ end lies upstream of two genes 5′ of ptxA (gvpA and gvpC [Fig. 4]). These 5′ genes encode gas vesicle proteins and are strongly upregulated during hormogonium differentiation (18). The 5′ region of gvpA is AT rich, and the putative −10 (AATACT) and −35 (TGTTAT) regions do not closely resemble a sigma 70-like factor recognition sequence (Fig. 4). The putative TSS is separated by 89 nucleotides from the translational start site. Because the genome location of the ptx genes most likely resulted from a duplication-insertion event and because these genes are naturally clustered (see Discussion), we refer to the ptx locus as inclusive of ptx genes, although the operon includes gvpAC.
FIG 4.
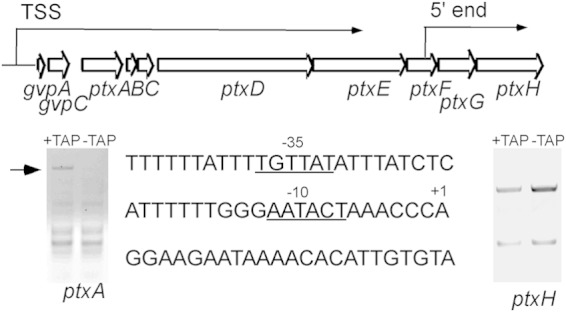
(Top) Locus 2i is drawn to scale, showing the approximate locations of the transcriptional start site (TSS) and the 5′ processed end of the mRNAs. (Bottom) (Left and right) Gels show the results of 5′ RACE. TAP, tobacco acid pyrophosphatase. (Center) Sequence of the putative promoter region.
To determine which genes in the locus may be essential for phototaxis function, mutations in various genes in locus 2i were constructed (Fig. 5). Mutants with an insertion mutation in ptxD (encoding an MCP homolog) or an in-frame deletion of ptxE (encoding a complete CheA homolog) were motile but nonphototactic. An insertion in ptxH resulted in a functional phototaxis phenotype, although the net movement toward light was less than that of the wild type (Fig. 5). This result indicates that this fragment of a CheA homolog is not essential for phototaxis function but may make a minor contribution. An insertion mutation in the upstream gvpA gene rendered the strain nonmotile. This result implies that an essential motility function is encoded not just in the hmp locus but also in the upstream genes of the ptx locus.
FIG 5.
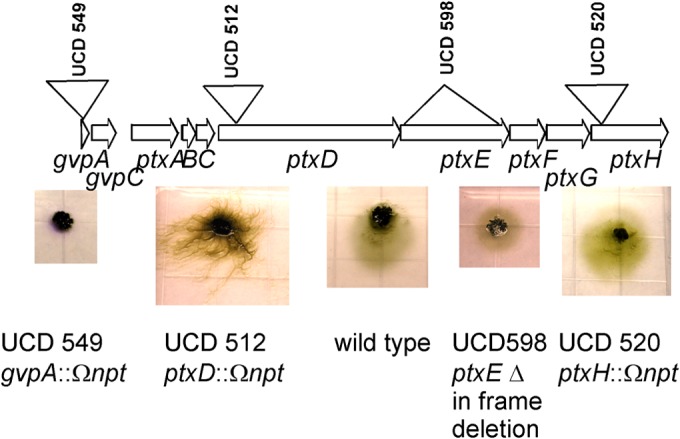
(Top) Locus 2i is drawn to scale, with upside-down triangles indicating the approximate locations of insertion mutations and right-side-up triangles indicating those of in-frame deletion mutations. (Bottom) Representative images of mutant strains after 24 h of incubation using white-light LEDs at 0.130 W/m2. An image of the wild-type strain is shown for comparison.
DISCUSSION
The results presented here document the phototactic behavior of N. punctiforme hormogonia. The motile filaments responded to white light at all intensities tested in a positive direction, and despite efforts at induction, N. punctiforme hormogonia did not exhibit high-light-intensity negative phototactic behavior as do the unicellular cyanobacterium Synechococus PCC 6803 (34) and vegetative filaments of Anabaena variabilis (35). Furthermore, the movement of cells toward white light was equivalent in distance and extent of response regardless of the intensity or flux of light, especially at low intensities (0.03 to 0.5 W/m2). This observation is consistent with the conclusion that the phototactic behavior is not directly linked to energy production or photosynthetic capacity; that is to say, it is not wholly a photokinetic response. The evidence presented here also establishes that a chromophore binding cyanobacteriochrome is required for the phototactic activity.
Bioinformatics identified 5 genetic loci in the N. punctiforme chromosome that contain genes encoding chemotaxis-like proteins that are likely to sense changes in environmental parameters and direct that information toward an output behavior. Our first exploration of these genes, through transcriptional analysis, revealed that all of them, except those in the alien locus L, were upregulated upon induction of the differentiation of hormogonia. It was not possible to discern, based purely on the patterns of transcriptional regulation, which genes were most relevant in phototaxis. Genes in two of the loci encode cyanobacteriochrome-MCPs, which should be primary candidates for direct phototaxis light receptors. Indeed, the full-length MCPs encoded in loci 2 and 2i are homologous to the concatenated PixJ1 and PixJ2 MCPs of Synechocystis PCC 6803 (21). However, it is equally possible that, for example, a cyanobacteriochrome-histidine kinase could initiate an indirect phosphorelay pathway that includes a nonphotoreceptor, taxis-like signal transduction system. We therefore employed mutant methodology and systematically inactivated a gene(s) within each of these chemotaxis-like loci to screen specifically for alterations in phototactic behavior. This avenue of inquiry revealed not only that locus 2i, encoding a cyanobacteriochrome-MCP, is the primary phototaxis locus but also that locus 1, encompassing the hmp genes, is essential for the synthesis of the motility mechanism (20) and that locus 3 affects hormogonium morphology. Deletion of the alien locus L resulted in no hormogonium morphological, motility, or phototactic defect, a finding consistent with the lack of transcriptional upregulation of the genes of this locus during hormogonium differentiation. In-frame deletion of the NpR6012 gene (pixJ) in locus 2, encoding a cyanobacteriochrome-MCP homolog of Synechocystis PCC 6803 PixJ1J2, also had no discernible effect on phototactic behavior or hormogonium morphology. This result is consistent with the obvious inability of this homologous pix locus to rescue the phototaxis-defective phenotype resulting from mutations in locus 2i.
Locus 2i genes, which we now term ptx genes, for phototaxis, are organized similarly to most of the other chemotaxis gene clusters of the cyanobacterial lineage. There are two genes encoding proteins with a CheY-like C-terminal response regulator receiver domain: ptxA encodes a PatA family protein, and ptxB encodes a typical CheY protein. The PatA family is characterized by an extended N-terminal domain whose sequence does not appear to be conserved across the genome. The PatA family CheY homolog (HmpA) in the hmp locus of N. punctiforme does not participate in phosphotransfer from the Hpt domain of HmpE, the colocated CheA-like protein (20). Moreover, hormogonia of the hmpA in-frame deletion mutant are motile, indicating that HmpA is not part of the Hmp signal transduction system for the activation of hps genes (20). This noncognate relationship may hold for the PatA family proteins in other cyanobacterial chemotaxis-like gene clusters as well.
In addition to potentially duplicated CheY proteins, other duplications are present in the ptx locus: ptxF encodes a second copy of CheW (ptxC encodes the first); ptxG encodes a partial copy of MCP (GAF and signaling domains); and ptxH encodes a partial copy of CheA (Hpt, histidine kinase, and CheW binding domains). To further dissect whether a gene is essential in phototaxis function, we again applied mutation genetics (Fig. 5). This proved to be a difficult process, because some of the genes in this locus are particularly difficult to mutate by gene replacement. In particular, however, the ptxE in-frame deletion indicates that no other CheA protein can substitute in the signaling complex. The nonmotile phenotype of the insertion mutation of the far-upstream gene gvpA (NpF2159) implies that the ptx locus encodes not only a phototaxis function but also functions that are critical to motility. We have not yet precisely pinpointed which of the three upstream ptx genes (ptxA, ptxB, or ptxC), or gvpA or gvpC, might be necessary for motility. Based on the precedent from the hmp locus, we hypothesize that the PatA family protein PtxA is not involved in phototaxis and could have other functions. A mutant with an alteration in ptxH (strain UCD 520), the last gene in the operon, retained phototactic behavior, although its movement was less than that of the wild type, indicating that at least this gene, and perhaps ptxG, which resides on the same processed mRNA, is not essential for a functional signal transduction complex.
Except for the alien locus, the CheA-like proteins in the five unicellular and filamentous cyanobacteria examined (21) form a monophyletic cluster, and those in N. punctiforme loci 2 and 2i appear to be paralogs. All of the CheA-like proteins in N. punctiforme, including that encoded by NpR0245 in alien locus L, contain a C-terminal response regulator receiver domain, and this appears to be the case for most, if not all, cyanobacteria. The C-terminal receiver domain is the preferred acceptor of phosphate from the Hpt domain of HmpE (20). Wuichet and Zhulin (21) suggested that locus 2i arose by duplication of locus 2. If so, it is likely that ptxF, ptxG, and ptxH arose by a second duplication event.
PtxD (encoded by NpF2164) differs from its homologs by the presence of 7 GAF domains, in contrast to 4 in PixJ of N. punctiforme (NpR6012) and Nostoc PCC 7120 (all1069) and 2 in Synechocystis PCC 6803 (sll0041). Each GAF domain can potentially bind a bilin (33); consequently, the photoresponse may be more integrated and more complex in those receptors with a higher number of GAF domains. Proteins homologous to PixJ and encoded by genes colocalized with other genes encoding chemotaxis-like proteins are present in 52 of 126 cyanobacterial genomes in the Integrated Microbial Genomes (IMG; DOE Joint Genome Institute) database (see reference 36). The cyanobacteriochrome domains contain 1 to 11 GAF domains. Except for those in Synechocystis PCC 6803 (37), Thermosynechococcus elongatus (38), Nostoc PCC 7120 (16), and N. punctiforme (39), cyanobacteriochrome domains in cyanobacterial strains have yet to be verified to bind a bilin chromophore and undergo reversible photoconversion. Only one other cyanobacterium, identified as “cyanobacterium PCC 7702,” contains a ptx locus encoding homologous proteins, with identical genome context (36; see also the IMG database). Cyanobacterium strain PCC 7702 was originally identified as a Chlorogloeopsis species, belonging to taxonomic subsection V (Stigonematales), whose members form hormogonia (40). It is now described as mostly unicellular, not differentiating hormogonia, and nonmotile (see supporting documents in reference 36). In cyanobacterium (Chlorogloeopsis) strain PCC 7702, the homologous ptxA gene is also preceded by the gvpA gene and the putative gvpC gene, and ptxF, ptxG, and ptxH homologs are present as the last three genes in the cluster.
The location of the transcriptional start site of the ptx locus is consistent with a duplication of the pix locus (locus 2) that did not include the original promoter region. The results of 5′ RACE analysis are also consistent with a single transcriptional start site for the operon. The gvpA promoter −10 (AATACT) and −35 (TGTTAT) sequences diverge from the consensus sigma 70 −10 recognition sequence (TANNNT) for cyanobacteria (41) primarily in that adenine replaces thymine at the 5′ end of the box. The promoters of many cyanobacterial genes have degenerate −35 sequences (41). Thus, the basis of upregulated transcription in N. punctiforme hormogonia and of the lack of expression in vegetative cells is not clear from the promoter sequence alone. Northern blot analysis (data not shown) of this region indicates that the mRNA is quite unstable, which is also implied by the multiple bands obtained in the ptxA primer extension gel (Fig. 4). This indicates that the message is short-lived and highly processed during hormogonium differentiation. A duplication event placing the promoterless ptx locus under the control of a gas vesicle gene promoter was fortuitous, since genes encoding gas vesicle proteins are expressed only in hormogonia (motile or not) by many of the cyanobacteria that differentiate hormogonia (4), including N. punctiforme (1).
ACKNOWLEDGMENTS
The initial experiments were supported by a grant from the National Science Foundation (IOS 0822008), and subsequent work was supported by an SISGR grant from the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DOE DE-FG02-09ER16117). R. Chen was supported by a fellowship from the China Scholarship Council.
We thank Delmar Larson for construction of the “black box” used for phototaxis assays and for assistance in acquiring the LED illumination system. Discussions with the SISGR group, especially Nathan Rockwell, were very productive. We also thank Becky Parales for helpful comments on the manuscript.
Footnotes
This publication is dedicated to Richard W. Castenholz, mentor and pioneer in studies of cyanobacterial motility.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02374-14.
REFERENCES
- 1.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111:1–61. [Google Scholar]
- 2.Castenholz RW. 1989. Oxyphotobacteria. Subsection IV. Order Nostocales, p 1780–1793. In Staley JT, Bryant MP, Pfennig N, Holt JG (ed), Bergey's manual of systematic bacteriology, 1st ed, vol 3 The Williams & Wilkins Co., Baltimore, MD. [Google Scholar]
- 3.Nultsch W, Schuchart H, Höhl M. 1979. Investigations on the phototactic orientation of Anabaena variabilis. Arch Microbiol 122:85–91. doi: 10.1007/BF00408050. [DOI] [Google Scholar]
- 4.Tandeau de Marsac N. 1994. Differentiation of hormogonia and relationships with other biological processes, p 825–842. In Bryant DA. (ed), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 5.Rippka R, Castenholz RW, Herdman M. 2001. Oxygenic photosynthetic bacteria. Subsection IV, p 562–589. In Boone DR, Castenholz RW, Garrity GM (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 1 Springer, New York, NY. [Google Scholar]
- 6.Choi JS, Chung YH, Moon YJ, Kim C, Watanabe M, Song PS, Joe CO, Bogorad L, Park YM. 1999. Photomovement of the gliding cyanobacterium Synechocystis sp. PCC 6803. Photochem Photobiol 70:95–102. [DOI] [PubMed] [Google Scholar]
- 7.Walsby AE. 1968. Mucilage secretion and the movements of blue-green algae. Protoplasma 65:223–238. doi: 10.1007/BF01666380. [DOI] [Google Scholar]
- 8.Hirose M. 1987. Energy supply system for the gliding movement of hormogonia of the cyanobacterium Nostoc cycadae. Plant Cell Physiol 28:587–597. [Google Scholar]
- 9.Castenholz RW. 1982. Motility and taxis, p 413–439. In Carr NG, Whitton BA (ed), The biology of cyanobacteria. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 10.Jékely G. 2009. Evolution of phototaxis. Philos Trans R Soc Lond B Biol Sci 364:2795–2808. doi: 10.1098/rstb.2009.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhaya D. 2004. Light matters: phototaxis and signal transduction in unicellular cyanobacteria. Mol Microbiol 53:745–754. doi: 10.1111/j.1365-2958.2004.04160.x. [DOI] [PubMed] [Google Scholar]
- 12.Bhaya D, Takahashi A, Grossman A. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc Natl Acad Sci U S A 98:7540–7545. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshihara S, Ikeuchi M. 2004. Phototactic motility in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Photochem Photobiol Sci 3:512–518. doi: 10.1039/B402320J. [DOI] [PubMed] [Google Scholar]
- 14.Ikeuchi M, Ishizuka T. 2008. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci 7:1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- 15.Yoshihara S, Suzuki F, Fujita H, Geng XX, Ikeuchi M. 2000. Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 41:1299–1304. doi: 10.1093/pcp/pce010. [DOI] [PubMed] [Google Scholar]
- 16.Narikawa R, Fukushima Y, Ishizuka T, Itoh S, Ikeuchi M. 2008. A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J Mol Biol 380:844–855. doi: 10.1016/j.jmb.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 17.Campbell EL, Meeks JC. 1989. Characteristics of hormogonia formation by symbiotic Nostoc spp. in response to the presence of Anthoceros punctatus or its extracellular products. Appl Environ Microbiol 55:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell EL, Christman H, Meeks JC. 2008. DNA microarray comparisons of plant factor- and nitrogen deprivation-induced hormogonia reveal decision-making transcriptional regulation patterns in Nostoc punctiforme. J Bacteriol 190:7382–7391. doi: 10.1128/JB.00990-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Risser DD, Meeks JC. 2013. Comparative transcriptomics with a motility-deficient mutant leads to identification of a novel polysaccharide secretion system in Nostoc punctiforme. Mol Microbiol 87:884–893. doi: 10.1111/mmi.12138. [DOI] [PubMed] [Google Scholar]
- 20.Risser DD, Chew WG, Meeks JC. 2014. Genetic characterization of the hmp locus, a chemotaxis-like gene cluster that regulates hormogonium development and motility in Nostoc punctiforme. Mol Microbiol 92:222–233. doi: 10.1111/mmi.12552. [DOI] [PubMed] [Google Scholar]
- 21.Wuichet K, Zhulin IB. 2003. Molecular evolution of sensory domains in cyanobacterial chemoreceptors. Trends Microbiol 11:200–203. doi: 10.1016/S0966-842X(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 22.Hoiczyk E, Baumeister W. 1998. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr Biol 8:1161–1168. doi: 10.1016/S0960-9822(07)00487-3. [DOI] [PubMed] [Google Scholar]
- 23.Duggan PS, Gottardello P, Adams DG. 2007. Molecular analysis of genes in Nostoc punctiforme involved in pilus biogenesis and plant infection. J Bacteriol 189:4547–4551. doi: 10.1128/JB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell EL, Summers ML, Christman H, Martin ME, Meeks JC. 2007. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J Bacteriol 189:5247–5256. doi: 10.1128/JB.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen MB, Arnon DI. 1955. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol 30:366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enderlin CS, Meeks JC. 1983. Pure culture and reconstitution of the Anthoceros-Nostoc symbiotic association. Planta 158:157–165. doi: 10.1007/BF00397709. [DOI] [PubMed] [Google Scholar]
- 27.Bensing BA, Meyer BJ, Dunny GN. 1996. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc Natl Acad Sci U S A 93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerhart E, Wagner H, Vogel J. 2008. Chapter 37. Approaches to identify novel non-messenger RNAs in bacteria and to investigate their biological functions: functional analysis of identified non-mRNAs, p 614–642. In Hartmann RK, Bindereif A, Schön A, Westhof E (ed), Handbook of RNA biochemistry. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 29.Cohen MF, Wallis JG, Campbell EL, Meeks JC. 1994. Transposon mutagenesis of Nostoc sp. strain ATCC 29133, a filamentous cyanobacterium with multiple cellular differentiation alternatives. Microbiology 140:3233–3240. doi: 10.1099/13500872-140-12-3233. [DOI] [PubMed] [Google Scholar]
- 30.Cai YP, Wolk CP. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol 172:3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen MF, Meeks JC. 1997. A hormogonium regulating locus, hrmUA, of the cyanobacterium Nostoc punctiforme strain ATCC 29133 and its response to an extract of a symbiotic plant partner Anthoceros punctatus. Mol Plant Microbe Interact 10:280–289. doi: 10.1094/MPMI.1997.10.2.280. [DOI] [PubMed] [Google Scholar]
- 32.Rockwell NC, Martin SS, Gan F, Bryant DA, Lagarias JC. 24 October 2014. NpR3784 is the prototype for a distinctive group of red/green cyanobacteriochromes using alternative Phe residues for photoproduct tuning. Photochem Photobiol Sci doi: 10.1039/C4PP00336E. [DOI] [PubMed] [Google Scholar]
- 33.Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. 2012. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry 51:1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- 34.Ng W-O, Grossman AR, Bhaya D. 2003. Multiple light inputs control phototaxis in Synechocystis sp. strain PCC6803. J Bacteriol 185:1599–1607. doi: 10.1128/JB.185.5.1599-1607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nultsch W, Mecke A. 1988. Photophobic responses of the cyanobacterium Anabaena variabilis. Arch Microbiol 150:343–347. doi: 10.1007/BF00408305. [DOI] [Google Scholar]
- 36.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, Calteau A, Cai F, Tandeau de Marsac N, Rippka R, Herdman M, Sivonen K, Coursin T, Laurent T, Goodwin L, Nolan M, Davenport KW, Han CS, Rubin EM, Eisen JA, Woyke T, Gugger M, Kerfeld CA. 2013. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci U S A 110:1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshihara S, Katayama M, Geng X, Ikeuchi M. 2004. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol 45:1729–1737. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]
- 38.Ishizuka T, Shimada T, Okajima K, Yoshihara S, Ochiai Y, Katayama M, Ikeuchi M. 2006. Characterization of cyanobacteriochrome TePixJ from a thermophilic cyanobacterium Thermosynechococcus elongatus strain BP-1. Plant Cell Physiol 47:1251–1261. doi: 10.1093/pcp/pcj095. [DOI] [PubMed] [Google Scholar]
- 39.Rockwell NC, Martin SS, Lagarias JC. 2012. Red/green cyanobacteriochromes: sensors of color and power. Biochemistry 51:9667–9677. doi: 10.1021/bi3013565. [DOI] [PubMed] [Google Scholar]
- 40.Rippka R, Herdman M. 2002. Pasteur Culture Collection of Cyanobacteria: catalogue and taxonomic handbook. I. Catalogue of strains. Institut Pasteur, Paris, France. [Google Scholar]
- 41.Curtis SE, Martin JA. 1994. Transcription in cyanobacteria, p 613–639. In Bryant DA. (ed), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]