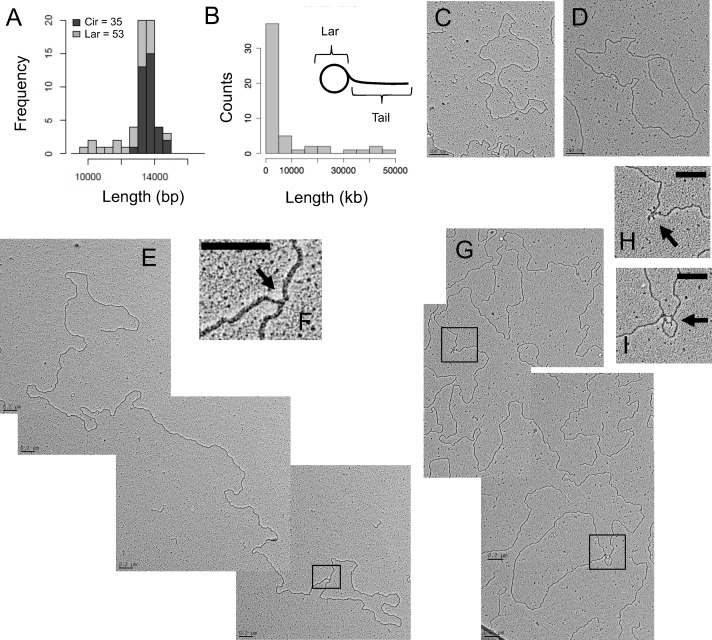
Fig 3. C. elegans mtDNA forms lariat-shaped rolling circle replication intermediates.
RNase I-treated mitochondrial nucleic acid was spread on parlodion-coated copper grids and visualized by TEM. (A) Frequency distribution of estimated strand-lengths of monomeric circular mtDNAs (Cir) and the circular portion of lariat mtDNAs (Lar) in kb. (B) Frequency distribution of estimated tail lengths of lariat mtDNAs, in kb. (C) Representative electron micrograph of a double-stranded DNA monomer circle. (D) Representative electron micrograph of an mtDNA lariat. (E) Multimeric lariat molecule with one genome unit-length circle (1n; 13.8 kb) with concatemeric linear tail (3.5n; 48.2 kb). (F) Higher magnification of the apparently double-stranded circle-tail junction. (G) Lariat with partial ssDNA structure consisting of a genome unit-length circle (1n; 13.8 kb) and concatemer tail (2.6n; 35.7 kb). Magnification of the single-stranded regions along the tail (H), as well as at the tail-circle junction (I); arrows indicate ssDNA. Scale bars, 0.2 μm. Data generated from three independent mtDNA isolations.