Figure 2.
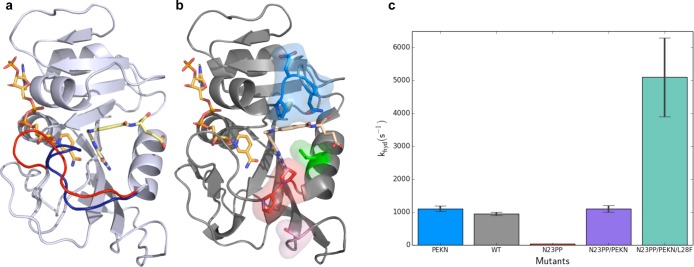
Dynamics in DHFR. (a) Crystal structures of E. coli DHFR show the Met20 loop in the occluded (1rx4, blue) and closed (1rx2, red) conformations. During the catalytic cycle of DHFR, this loop fluctuates between these conformations on the millisecond time scale. The ligands NADPH (left ligand) and folate (right ligand) are shown in orange and yellow, respectively. (b) Mutation of Asn23 to two proline residues (N23PP) shown as sticks in red and Ser148 to alanine (S148A) shown in pink reduce activity of ecDHFR. Mutation of Gly51 to the sequence PEKN (shown in blue) partially recovers the catalytic activity. The activity is increased further by the Leu28Phe (L28F, green) mutation. The structure of N23PP/PEKN (4gh8) is shown with NADPH shown in orange, with substrate mimic methotrexate in tan. (c) pH independent hydride transfer rates of different mutants show the quantitative effects of mutations that alter the dynamics of the Met20 loop and packing around the substrate.
