Figure 3.
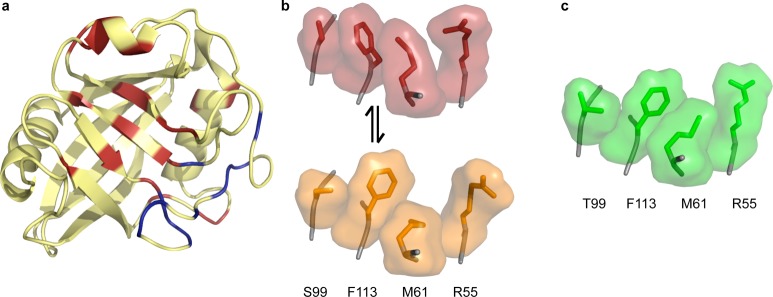
Dynamics in CypA. (a) Exchanging residues detected by CPMG experiments show two groups with exchange rate kex = 1140 ± 200 s–1 (red) and kex = 2260 ± 200 s–1 (blue) in the absence of substrate. All residues with detectable dynamic exchange can be fit to one rate (∼2400 s–1) when the protein is saturated with substrate, which interconverts from cis to trans on the enzyme (1rmh). (b) Wild-type CypA (3k0n) shows two sets of conformations at room temperature. The network of side chains of residues S99, F113, M61, and R55 are shown with surface representations around sticks, with the major conformation in red and the minor conformation in orange. These residues lie across the central β strands shown in panel a. (c) The network of these four residues for the S99T mutant at room temperature only occupies the minor-like conformation, shown in green (3k0o).
