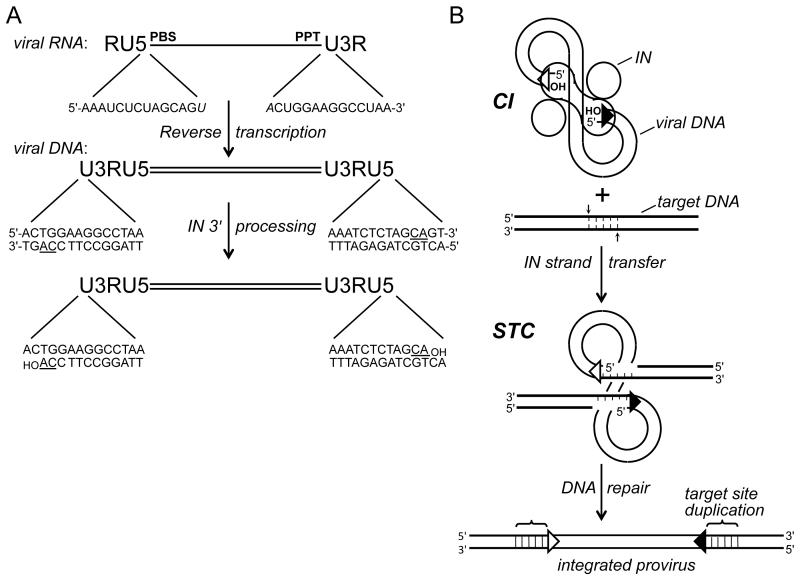
Figure 1.
Integration substrate and integrase activities. (A) Reverse transcription yields linear, double-stranded DNA with U3RU5 long terminal repeats. The 14 terminal bp of HIV-1 DNA are compared with their originating positions in viral RNA (italicized bases form part of the primer binding site (PBS) and polypurine tract (PPT) that are important for DNA synthesis). The positions of the invariant CA dinucleotides (underlined in the DNA) relative to the 3′ ends of the PBS and PPT determines whether 3′ processing is required to yield a CAOH 3′ terminus; 3′ processing by HIV-1 integrase liberates a pGpTOH dinucleotide from each viral DNA end. (B) Two monomers (oblong shape) of an HIV-1 integrase tetramer within the cleaved intasome (CI) use the viral DNA 3′-hydroxyl groups to cut the target DNA (thick bold lines) with a 5 bp stagger, which concomitantly joins the LTR ends to target DNA 5′ phosphates. Repair of the strand transfer complex (STC) yields a 5 bp duplication of target DNA flanking the integrated HIV-1 provirus. Open and filled triangles, U3 and U5 termini. Integrase was omitted from the drawing of the STC for simplicity. IN, integrase.