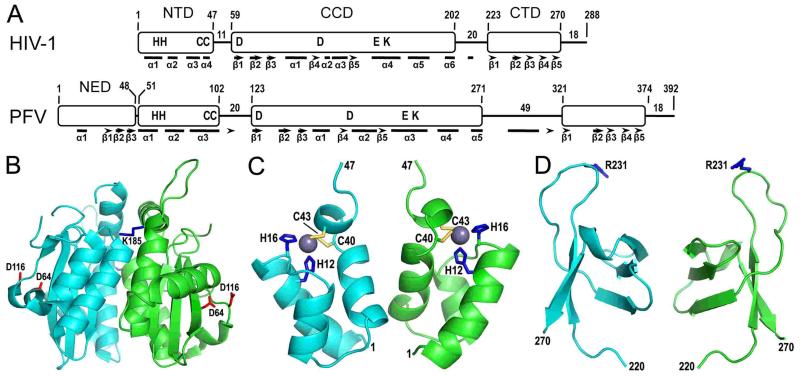
Figure 2.
Retroviral integrase domain organization and HIV-1 integrase domain structures. (A) The N-terminal domain (NTD), catalytic core domain (CCD), and C-terminal domain (CTD) are common among all retroviral integrase proteins, whereas sequence analysis indicates that gammaretrovirus and epsilonretrovirus in addition to spumavirus proteins harbor an N-terminal extension domain (NED) (55, 94). HIV-1 and PFV integrases were aligned by NTD N-termini, with positions of domain boundaries and lengths of interdomain linker and C-terminal tail regions indicated. Residues conserved across all retroviral integrase proteins are shown in single letter code. Bars and arrows indicate alpha helix and beta strand secondary elements as determined by X-ray crystallography for PFV integrase (94) and by a combination of X-ray crystallography (66, 149) and molecular modeling (105) for HIV-1 integrase. (B) The X-ray crystal structure of the HIV-1 integrase CCD [protein database (pdb) code 1ITG] (43) highlights in red sticks the aspartate residues of the DDE catalytic triad (the glutamic acid was not visualized in this structure) and in blue sticks the Lys185 substitution that enhanced protein solubility and enabled protein crystallization (the Lys residue at the rear face of the dimer is barely visible in this projection). (C) The NMR structure of the integrase NTD (pdb code 1WJC) highlights the His (blue sticks) and Cys (yellow sticks) residues that coordinate a single zinc atom (grey sphere) (99). (D) The structure of the HIV-1 integrase CTD as determined by NMR (pdb code 1IHV) (142) highlights Arg231, which has been implicated in target DNA binding (108, 166).