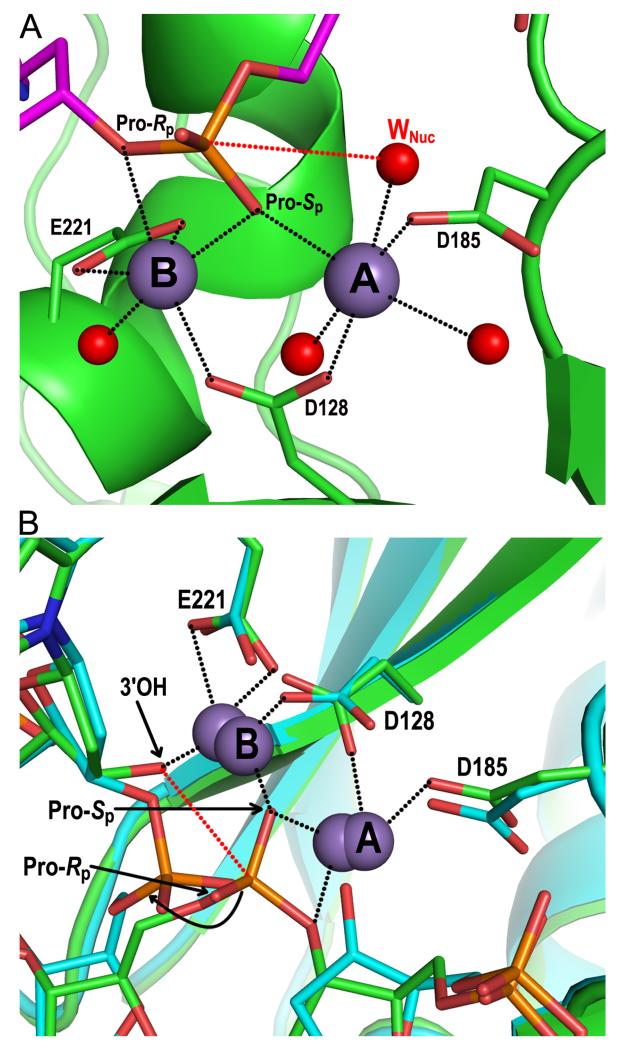
Figure 6.
Structural basis of integrase 3′ processing and strand transfer activities. (A) Structure of the manganese-bound SSC (pdb code 4E7I). The DNA and integrase backbones are colored magenta and green, respectively; red and orange sticks are oxygen and phosphorus atoms, respectively. Gray and red spheres are manganese ions and water molecules, respectively, with the nucleophilic water labeled WNuc. Black dashed lines indicate metal ion interactions; the red dashed line connects the nucleophile and scissile phosphodiester bond. (B) Overlay of metal ion-bound TCC (pdb code 4E7K; DNA and protein in green) and STC (pdb code 4E7L; elements painted in cyan) structures. Both sets of metal ions are shown; the 3.8 Å spacing between ions in the TCC contracts to 3.2 Å in the STC (85). The curved black line indicates the displacement of the viral-target DNA phosphodiester bond after strand transfer relative to the scissile bond in target DNA. Other labeling is the same as in panel A.