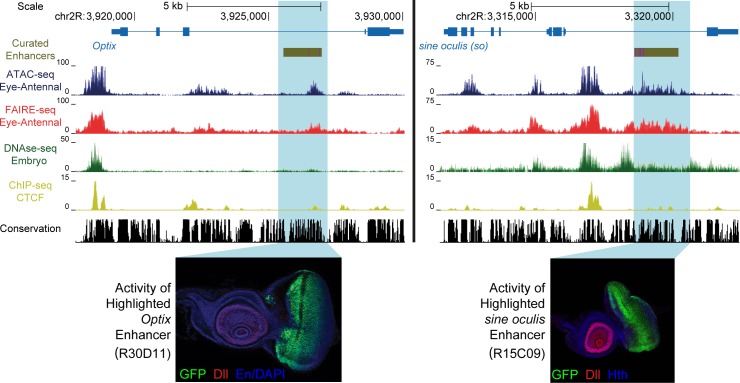
Fig 1. Enhancer identification in the eye primordium by ATAC-seq and FAIRE-seq.
Two example eye-related genes from the Drosophila melanogaster genome (dm3). Tracks shown include: experimentally validated enhancers from ORegAnno and REDFly [72,73] (shown in Gold), coverage data in the eye-antennal disc for ATAC-seq (Blue), FAIRE-seq (Red) and CTCF ChIP (Yellow), DNaseI-seq (Green) from whole stage 11 embryos [8] and finally sequence conservation across 14 Drosophila species (Black). On the left the gene locus of Optix is shown. The region in gold is a known enhancer with binding sites for Eyeless overlaid in purple, with FlyLight enhancer activity of R30D11 below. This region is identified as a peak in both ATAC-seq and FAIRE-seq in the eye, but not in embryonic DNaseI-seq. On the right is the gene locus of sine oculis (so). The region in gold is a known enhancer with binding sites for Eyeless and Twin of Eyeless overlaid in purple. The reporter activity of this enhancer (R15C09) is shown by the Janelia Farm FlyLight project. Again, this enhancer is identified as a peak in both ATAC-seq and FAIRE-seq but not in embryonic DNaseI-seq. In both examples, peaks can be seen in the CTCF ChIP-seq data corresponding to regions identified as open by all three other techniques. Some small differences between ATAC-seq and FAIRE-seq can also be seen, including a higher background signal for FAIRE-seq. Images obtained from the FlyLight database (http://flweb.janelia.org/cgi-bin/flew.cgi).