Figure 2.
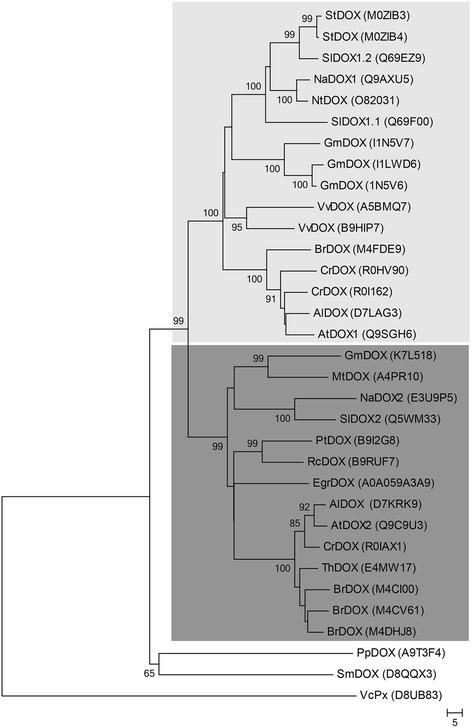
Phylogenetic tree of confirmed and putative α-dioxygenases. Full-length amino acid sequences of available and putative α-DOX proteins were aligned by CLUSTAL W and a phylogenetic tree was constructed by the neighbor-joining method using MEGA version 5.05. Numbers at branch nodes represent the confidence level of 1000 bootstrap replications. The identities of the individual α-DOX protein sequences are indicated by their uniprot entry number (http://www.uniprot.org). Two clusters are highlighted in the phylogenetic tree including α-DOX1 (light grey) and α-DOX2 proteins (dark gray) respectively. The abbreviations of species are as follows: Al: Arabidopsis lyrata, At: Arabidopsis thaliana, Br: Brassica rapa, Cr: Capsella rubella, Eg: Eucalyptus grandii, Gm: Glycine max, Mt: Medicago truncatula, Na: Nicotiana attenuata, Nt: Nicotiana tabacum, Pp: Physcomitrella patens, Pt: Populus trichocarpa, Rc: Ricinus communis, Sl: Solanum lycopersicum, Sm: Selaginella moellendorffii, St: Solanum tuberosum, Th: Thellungiella halophila, Vc: Volvox carteri, Vv: Vitis vinifera.
