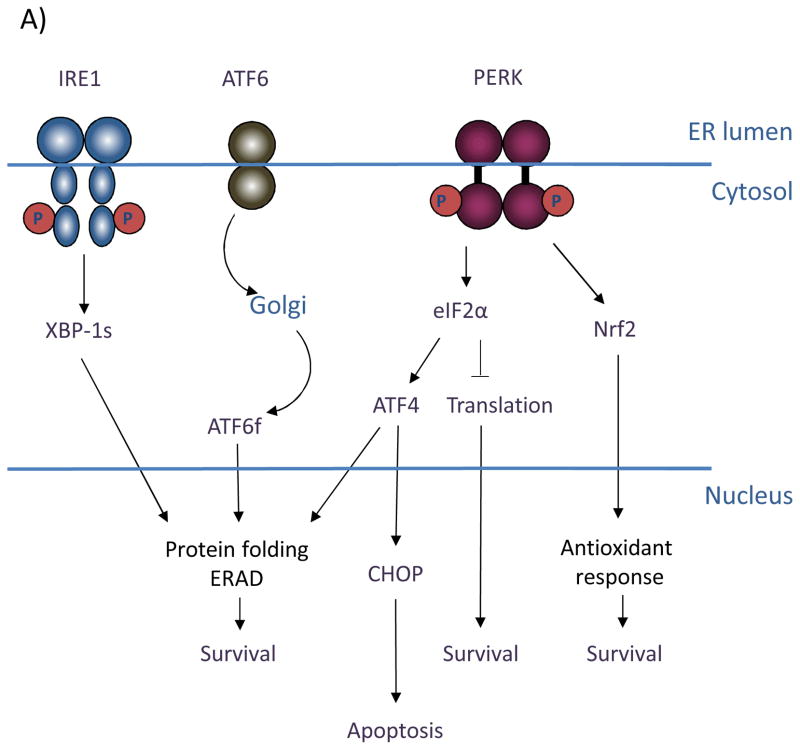
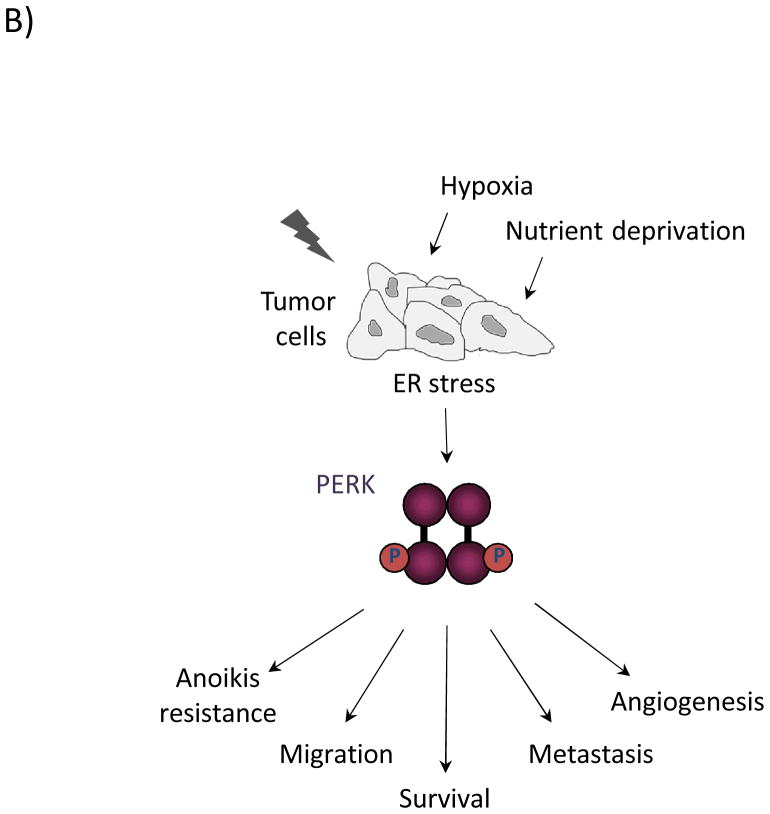
Figure 1. (A) Signaling through the three branches of the Unfolded Protein Response.
The ER stress sensors inositol-requiring gene 1 (IRE1), activating transcription factor 6 (ATF6), and (PKR)-like ER kinase (PERK) span the ER membrane. In response to conditions that perturb ER homeostasis, IRE1 is activated through oligomerization and trans-autophosphorylation. IRE1 RNase activity cleaves the mRNA of XBP-1 to generate an active transcription factor (XBP-1s) that targets genes involved in protein folding and ER-associated degradation to facilitate ER stress resolution and ultimately promote cell survival. Activation of ATF6 in response to ER stress involves its migration to the trans-Golgi, where it is proteolytically processed. This cleavage event releases the cytosolic bZIP domain, which translocates to the nucleus to activate the expression of ER chaperones and ERAD components. PERK activation occurs through oligomerization and trans-autophosphorylation, leading to phosphorylation of the translation initiation factor, eIF2α, and the antioxidant response factor, Nrf2. Phosphorylation of eIF2α leads to general inhibition of translation, contributing to overall cell survival. Phosphorylation of Nrf2 activates transcription of antioxidant factors. Selective upregulation of the bZIP transcription factor ATF4 through eIF2α targets both pro-survival genes as well as the pro-apoptotic factor GADD153/CHOP. (B) Oncogenic functions of PERK. Restricted nutrient and oxygen conditions in the tumor microenvironment triggers UPR signaling in cancer cells. The cell adaptive nature of PERK signaling enables enhanced cell survival, increased migratory and metastatic capacity, resistance to anoikis in ECM-detached cells, and increased pro-angiogenic potential to support tumor growth.