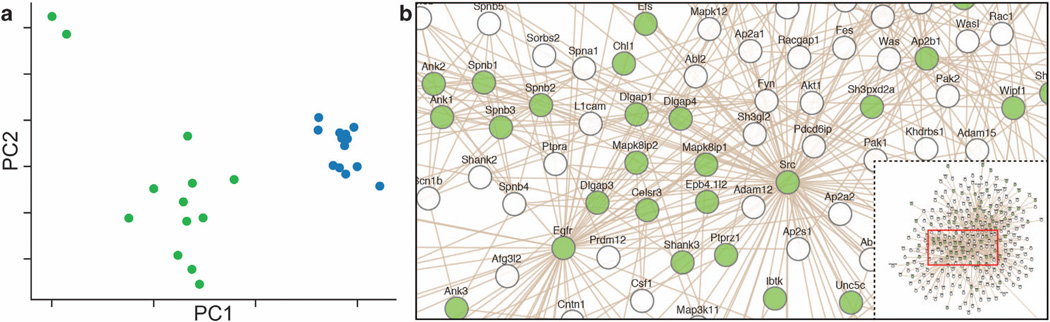
Figure 1.
(a) Principal component analysis of expression profiles from paired synaptoneurosome (SN) and total homogenate (TH) samples are shown in green and blue, respectively. Preparation difference is the first principal component and explains 17% of the variance. (b) SN-enriched mRNA network illustrating known protein interactions between the SN-enriched mRNAs. The bottom right portion of the figure is an overview of the entire network, and the highlighted portion of this network has been enlarged. The green nodes represent the mRNAs enriched in the SN compared with the TH preparation (fold-change threshold of > 10%, Benjamini–Hochberg method P < 0.05). Many known synaptic mRNAs are found in the center of this network, emphasizing enrichment of the synaptic components in the SN preparation.