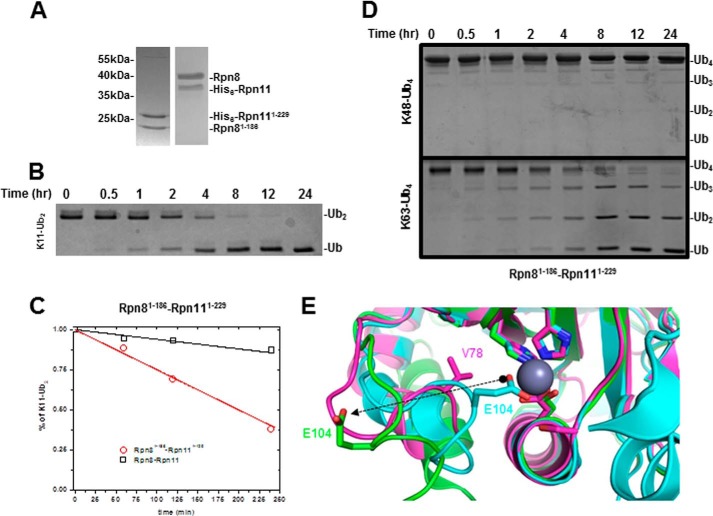
FIGURE 7.
A, SDS-PAGE comparing truncated Rpn8(1–186)·Rpn11(1–229) and wild type heterodimer. B, DUB activity of truncated heterodimer was assessed on Lys11-Ub2. C, initial processing of Lys11-Ub2 plotted for truncated (red) and wild-type heterodimer (black). D, truncated heterodimer fails to process Lys48-Ub4 efficiently (top); however, it is competent to process Lys63-Ub4. E, structural alignment of full-length CSN5 (Protein Data Bank entry 4D10; cyan), truncated CSN5(1–257) (Protein Data Bank entry 4F7O; green), and truncated Rpn11(2–239) (Protein Data Bank entry 4O8X; purple) shows classical MPN motif coordinating the active site Zn2+ ion. Glu104 (sticks) undergoes a 12-Å transition between the two forms of CSN5, regulating access to the Zn2+ ion in the process. Val78 (purple sticks) of Rpn11 is the equivalent of Glu104 in CSN5.