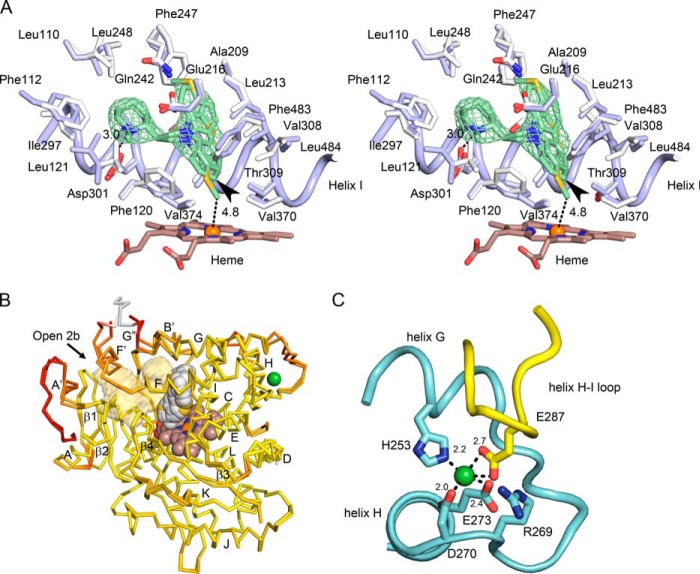
FIGURE 2.
Structural features of the closed P450 2D6 thioridazine complex crystallized in the C121 space group. Oxygen, nitrogen, sulfur, iron, and nickel atoms are colored red, blue, yellow, orange, and green, respectively. A, a stereo view (cross-eyed) of the active site with carbons colored blue for the thioridazine complex and white for the 3QM4 structure of the P450 2D6 prinomastat complex for comparison of changes in side chains and backbone atoms (Cα). The heme is depicted as a stick figure with carbons colored brown. The green mesh depicts a 2m|Fo| − D|Fc| ligand omit map contoured at 1 σ around thioridazine. The distance of the thioridazine atom that most closely approaches the heme iron is depicted as a dashed line with the length indicated in Å. C7 of thioridazine is indicated by the arrowhead. B, an overlay of the two chains in the asymmetric unit colored to reflect variation of root mean square deviation for Cα atoms from low (yellow) to high (red). The alignment and color map were generated by ProSMART. The heme (brown carbons), thioridazine (gray carbons), and a nickel (green) are rendered as spheres. The antechamber adjacent to the active site is rendered as a transparent light orange surface. The arrow indicates the entrance to channel 2b. Helices are denoted by letters and sheets by numerals as customary for P450s. C, a view of the nickel ion binding site in chain B (cyan carbons) and the interaction of Glu-287 in a neighboring copy of chain B (yellow carbons) with the nickel ion. Distances between oxygen and nitrogen atoms forming the nickel binding site are indicated in Å.