Figure 1.
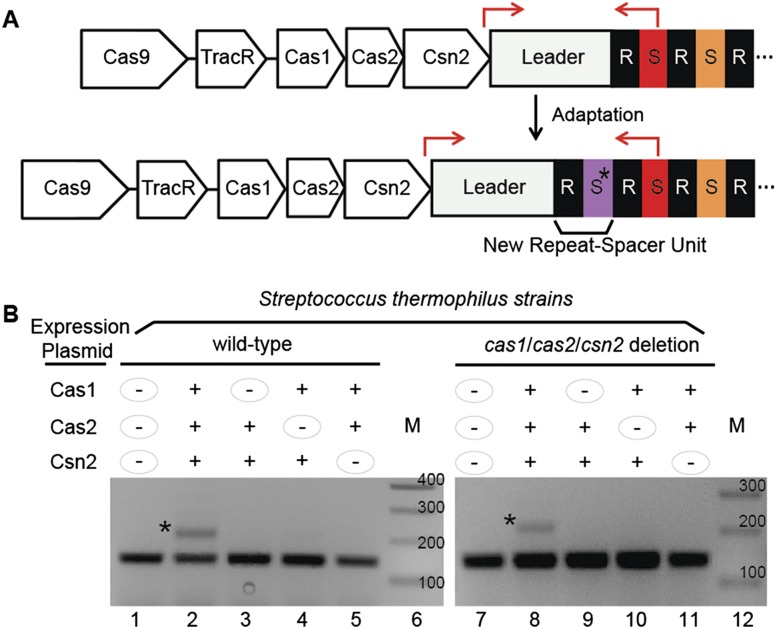
Cas1, Cas2, and Csn2 are critical for adaptation. (A) Illustration of CRISPR1 module and adaptation. (Top) The S. thermophilus DGCC7710 CRISPR1 module encodes four Cas proteins: Cas1, Cas2, Csn2, and Cas9. The tracrRNA is encoded between Cas9 and Cas1. The CRISPR leader sequence (gray) is followed by 32 repeat (R, black)–spacer (S, colored) units (only a few of which are shown for simplicity). The locations of primers used in PCR for detection of new spacer acquisition are indicated by red arrows. (Bottom) The product of acquisition of a novel spacer (asterisk; purple) and an associated repeat is illustrated. (B) Analysis of spacer acquisition in wild-type (left) and cas1/cas2/csn2 deletion (right) strains expressing combinations of Cas1, Cas2, and Csn2 proteins from plasmids. The leader-proximal region of CRISPR1 was PCR-amplified with primers indicated in A. Plasmid expression of individual proteins is indicated with “+” or “−” signs. The PCR product corresponding to addition of one repeat–spacer unit is indicated with an asterisk. Sizes of DNA standards (M) are indicated. Data representative of multiple experiments are shown.