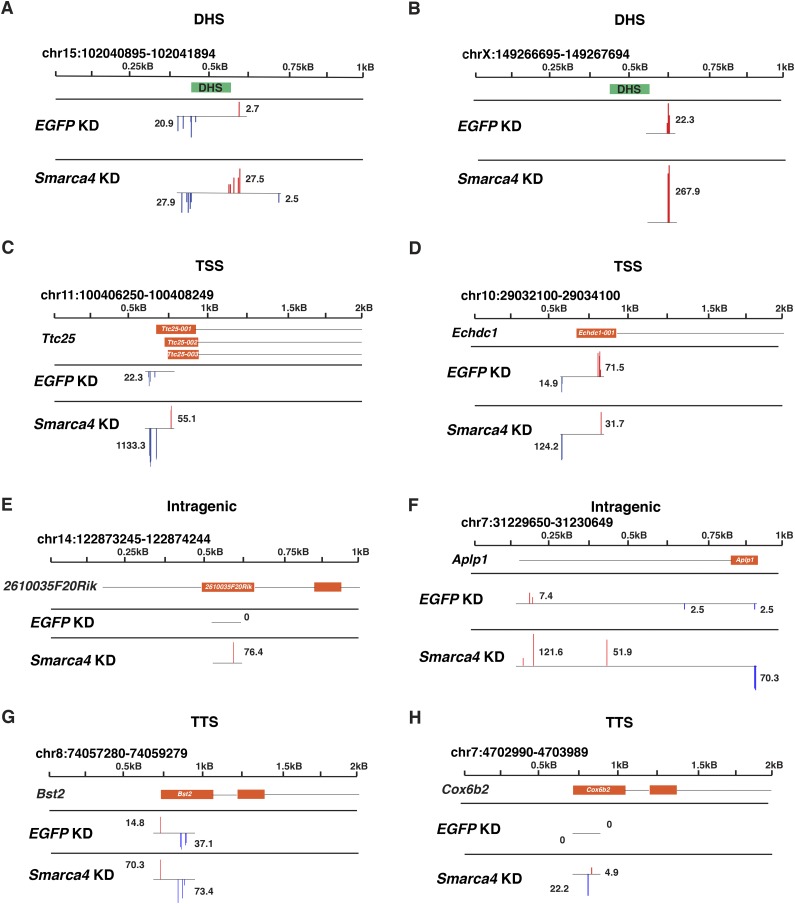
Figure 1.
The esBAF complex inhibits production of noncoding transcripts initiating from multiple genomic features. Genome browser tracks of two DHSs (A,B), two TSSs (C,D), two intragenic regions (E,F), and two TTSs (G,H). Bars corresponding to normalized CapSeq reads from EGFP knockdown (KD) and Smarca4 knockdown are shown in log2 scale at their initiation sites, and the (real space) number of normalized reads per cluster of transcripts is noted in each track. Browser tracks of normalized CapSeq reads of one replicate from EGFP knockdown and Smarca4 knockdown are shown. Isoforms are shown in an orange box below the scale, with introns indicated as black lines. DHSs are indicated in green boxes. Blue bars indicate transcription from the Crick strand, while red bars indicate transcription from the Watson strand. It should be noted that transcripts near TSSs in the same orientation as the coding sequence likely depict mRNAs not ncRNAs.