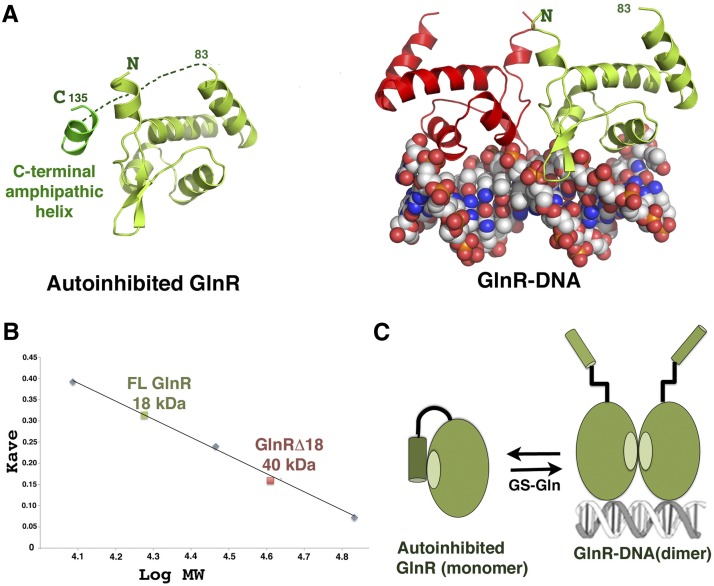
Figure 3.
Molecular mechanism of GlnR autoinhibition. (A, left) Structure of apo GlnR with proposed autoinhibitory contacts present in the crystal structure. (Right) The DNA-bound form showing the subunit in the same orientation. (B) SEC of FL GlnR and GlnRΔ18 at 3 mg/mL showing that FL GlnR is a monomer and that GlnRΔ18 is a dimer. (C) Schematic model for GlnR autoinhibition and activation by GS. The C-terminal region of GlnR folds back and forms an autoinhibitory helix that inhibits dimer formation. Binding of this C-terminal tail by GS–Gln relieves autoinhibition, shifting the equilibrium to the DNA-binding active form.