Fig. 5.
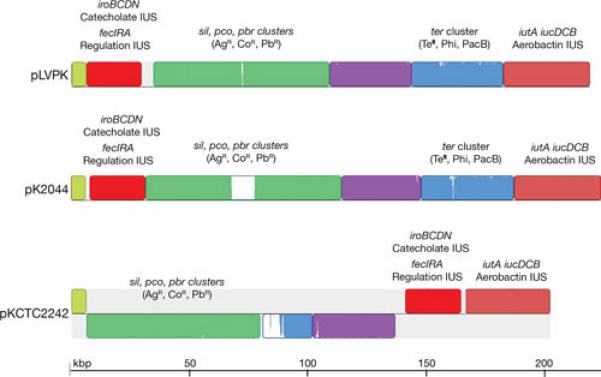
Multiple alignment of pLVPK, pK2044 and pKCTC2242. The nucleotide sequences of pLVPK (accession number AY378100.1)(112), pK2044 (accession number AP006726.1)(116), and pKCTC2242 (accession number CP002911.1)(117) were compared using the MAUVE aligner version 2.3.1 (149). Different colors represent local LCBs. Inside each block there is a similarity profile of the sequence, the height corresponds to the average level of conservation. Completely white areas are not aligned and most probably contain sequences specific to the particular molecule. In pKCTC2242 the LCBs drawn below the black line are inverted with respect to their homologs in pLVPK and pK2044. Some genes or clusters present in these blocks are identified by name. The terZ gene has been reported as “truncated” (112). The truncation is a consequence of an extra T in the sequence that could also be a sequencing error. The terBCDE genes are sufficient for the tellurite resistance phenotype (TeR). The ter cluster is also responsible for the phage inhibition (Phi) and colicin resistance (PacB) phenotypes (150). Copper (pco), silver (sil), lead (pbr), and tellurite (ter) resistance related genes; IUS, iron uptake system.
