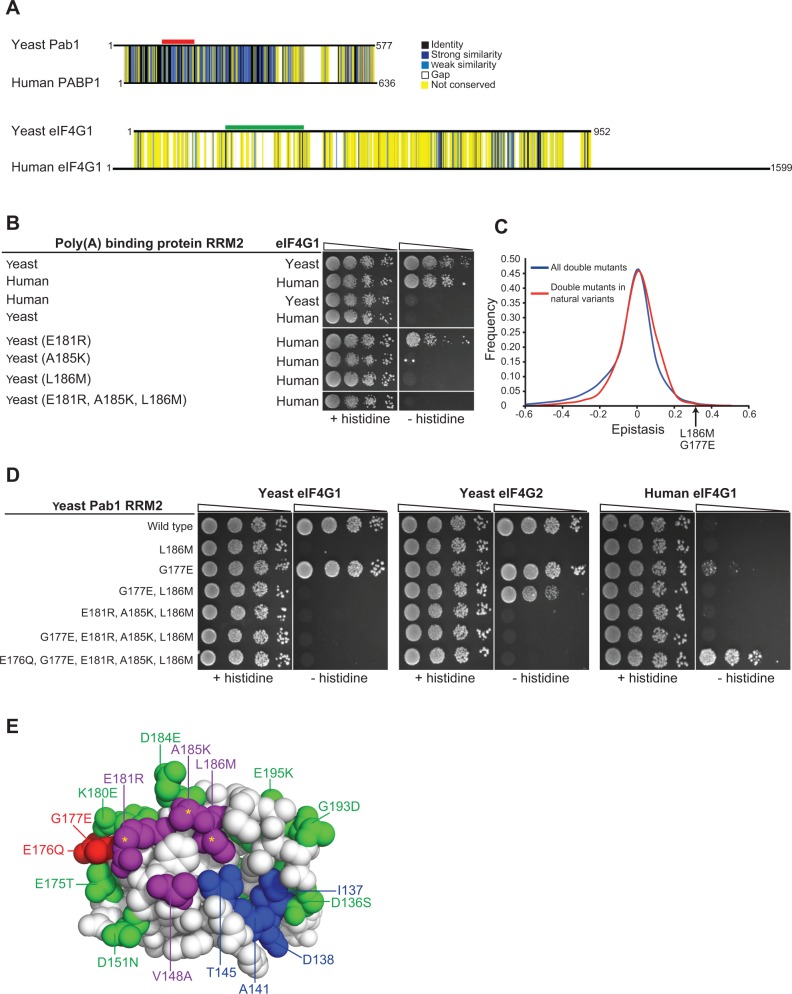
Figure 6. Testing humanizing substitutions for their ability to change the binding specificity of the yeast Pab1 RRM2 to the human eIF4G1.
A) Pairwise alignments of the yeast and the human orthologues of the poly(A)-binding protein-1 and eIF4G1 were generated using Clustal Omega [43]. Black horizontal lines designate the protein sequences. Degrees of similarity between aligned positions as well as gaps are color coded with the color key depicted on the left. The yeast Pab1 sequence that is associated with eIF4G1 binding [22] is highlighted with a red line and the yeast eIF4G1 region associated with Pab1 binding [38] is shown with green. B) Yeast two-hybrid assays testing Pab1 RRM2 variants that carry the specified mutations for their ability to interact with the N-terminus (amino acids 1–260) of the human eIF4G1. Cells were serially diluted and seeded as described in Fig. 2. C) Epistasis score distributions of Pab1 variants carrying two mutations that are either found together in natural homologues of the poly(A)-binding protein (red line) or not (blue line). A positive epistasis score indicates a growth rate of the double mutant that was better than expected based on the effects of each of the constituent single mutants alone. An arrow shows the epistasis score of the G177E and L186M double mutant. D) Yeast two-hybrid assays testing Pab1 RRM2 variants that carry the specified mutations for their ability to interact with the N-termini of the yeast eIF4G1 and eIF4G1 and with the human eIF4G1. E) An upper view of the eIF4G1 contact surface of the human PABP1 RRM2 domain (PDB ID 1CVJ). Differences between the yeast and the human sequences are specified by the amino acid substitution nomenclature. eIF4G1 contact site residues that are conserved between yeast and human are shown in blue and contact site residues that differ between the two species are in purple. Deleterious substitutions are marked with yellow asterisks and non-deleterious substitutions are colored in green. E176Q and G177E are shown in red. All other conserved residues between yeast and human are white.