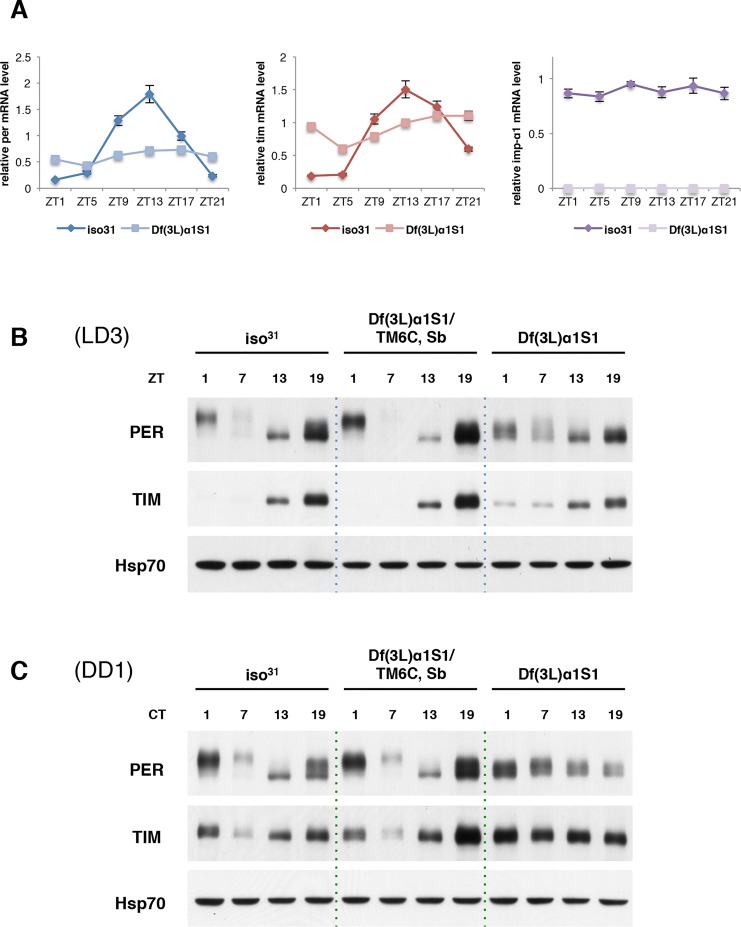
Figure 3. Molecular oscillations of PER and TIM are dampened in Df(3L)α1S1 flies.
(A) Quantitive PCR (qPCR) reveals that per and tim mRNA oscillations are blunted in Df(3L)α1S1 flies relative to those in wild-type flies. importin α1 mRNA levels do not cycle. actin was used as an internal control to normalize transcript levels. The quantification curves in each panel were plotted as average ± standard error of the mean (SEM) of three independent experiments. (B and C) Western blots of adult fly heads of wild-type (iso31), heterozygous, and homozygous Df(3L)α1S1 flies were probed for PER and TIM. Flies of the indicated genotypes were collected (B) at different zeitgeber times (ZT) on the 3rd day of LD (LD3) and (C) at different circadian times (CT) on the 1st day of DD (DD1). The blots were also probed with antibody to Hsp70 as a loading control. Similar results were obtained in two or three independent experiments. The quantification of TIM expression levels is shown in S5 Fig.