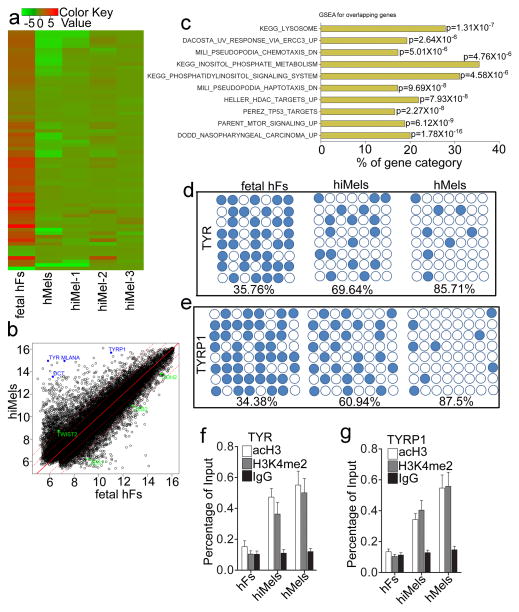
Figure 7. Molecular characterization of induced human melanocytes.
a. Heat-map of genes differentially expressed in RNA-microarray analysis performed on human fetal fibroblasts (fetal hFs), induced melanocytes derived from human fetal fibroblast (hiMels) and normal skin melanocytea (hMels). b. Scatter plots show that melanocytic markers are expressed in hiMels, but not in fetal hFs. c. Gene Set Enrichment Analysis (GSEA) for the overlapping genes between hMels and hiMels. Many gene sets including KEGG_LYSOSOME, DACOSTA_UV_RESPONSE_VIA_ERCC3, PARENT_MTOR_SIGNALING_UP, MILI_PSEUDOPODIA_HAPTOTAXIS_DN and MILI_PSEUDOPODIA_CHEMOTAXIS_DN were enriched in hiMels and hMels. d and e. DNA methylation analysis of the promoters of TYR (d) and TYRP1 (e) in fetal hFs, hiMels and hMels. Open circles indicate unmethylated CpG dinucleotides, while closed circles indicate methylated CpGs. f and g. Histone modification analysis of promoters of TYR and TYRP1 in fetal hFs, hiMels and hMels. Chromatin immunoprecipitation was performed using antibodies against dimethylated histone H3K4 (H3K4me2) and H3 acetylation (acH3). TYR and TYRP1 promoters showed enrichment for the active states (H3K4 me2 and acH3) in hiMels and hMels. In fetal hFs, TYR and TYRP1 promoters appeared in the inactive state. Representative data are from three independent experiments.