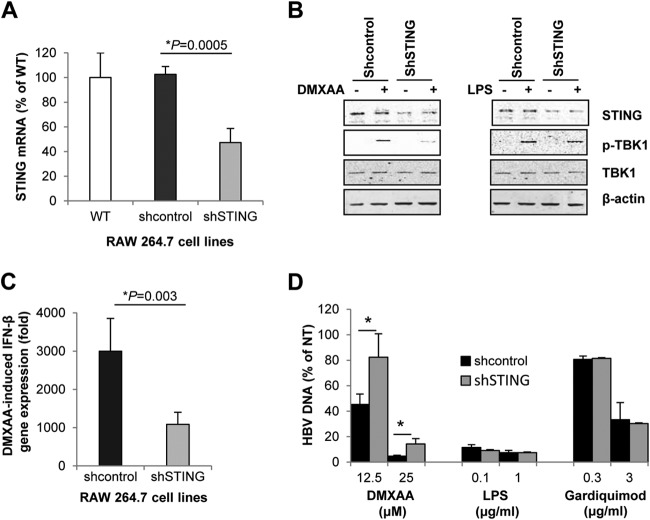
FIG 6.
DMXAA-induced cytokine and antiviral responses were STING dependent. (A) Total RNAs were extracted from parental wild-type (WT) RAW264.7 cells and RAW264.7-derived stable cell lines that express scrambled shRNA (shcontrol) or shRNA targeting mouse STING mRNA (shSTING). STING mRNA levels were determined by real-time RT-PCR, and data (mean values ± standard deviations; n = 3) are expressed as percentages of the STING mRNA in wild-type RAW264.7 cells. (B) RAW264.7 cells expressing shcontrol or shSTING were mock treated or treated with 125 μM DMXAA for 30 min. Expression and activation of STING and TBK1 were determined by Western blot assays. Cells treated with 1 μg/ml of LPS served as controls. β-Actin served as a loading control. (C) Knockdown of STING abrogated DMXAA-induced IFN-β gene expression. RAW264.7-derived shcontrol and shSTING cells were treated with 125 μM DMXAA for 3 h. IFN-β mRNA was quantified by a real-time RT-PCR assay. Data (mean values ± standard deviations; n = 3) are expressed as fold induction of gene expression relative to that in untreated controls. (D) Knockdown of STING compromised DMXAA-induced antiviral activity. AML12HBV10 cells cultured in the absence of tetracycline for 1 day were mock treated or treated for 2 days with 50% of the conditioned media harvested from RAW264.7-derived shcontrol or shSTING cells, which were treated for 12 h with the concentrations of DMXAA, LPS, or Gardiquimod indicated. Cytoplasmic HBV core DNA was extracted and quantified by a real-time PCR assay. AML12HBV10 cells treated with conditioned media from mock-treated RAW267.4 cells served as a control (NT). HBV DNA levels (mean values ± standard deviations; n = 3) are expressed as percentages of the NT control value. Data were statistically analyzed by Student's t test. *, P < 0.05.