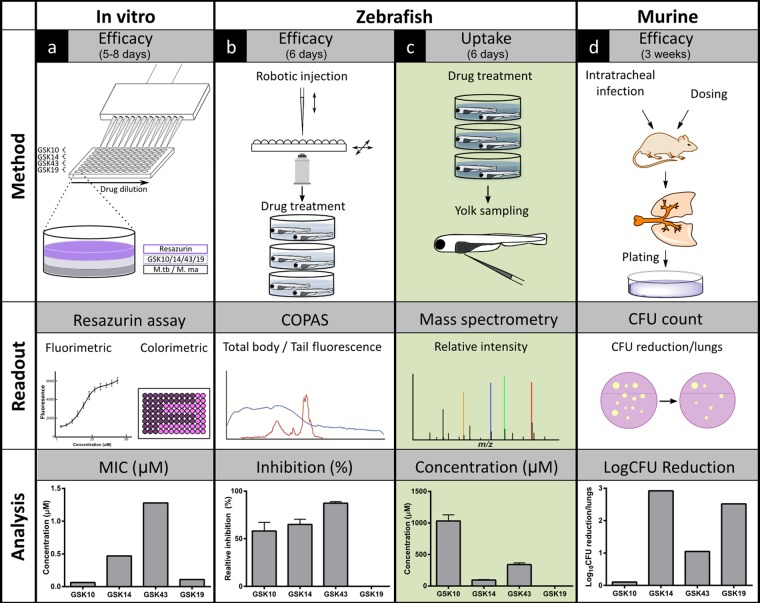
FIG 1.
Scheme of drug efficacy methods using antibacterial activity and uptake assays in the zebrafish model integrated in conventional disease-screening pipeline. Different models are used in our workflow to determine the efficacy of antitubercular compounds (from left to right). (a) Initially, the in vitro efficacy of the tested compounds is determined by their MIC against Mycobacterium marinum and Mycobacterium tuberculosis cultures using fluorimetric and colorimetric readouts from the resazurin assay. (b to d) Biological validation subsequently is performed in in vivo models. (b) First, compound efficacy is screened in M. marinum-infected zebrafish larvae. Embryos are robotically injected, and following compound treatment, the percentage of inhibition is determined using the fluorescence readout of the COPAS system. (c) To unravel whether certain compounds fail to be active in zebrafish larvae due to the lack of antibacterial activity or poor uptake, our microneedle sampling method combined with mass spectrometry is used to assess uptake levels from samples of the yolk. (d) As a gold standard in antitubercular drug development, compound efficacy is established by determining the rate of the CFU reduction in the lungs of M. tuberculosis-infected rodents. After setting (arbitrary) cutoffs in all models, compounds could be categorized into positive or negative groups. By the comparison of these groups along the pipeline, our improved zebrafish platform may give a more predictive value for human efficacy of drugs. Cycle times are indicated for each model.