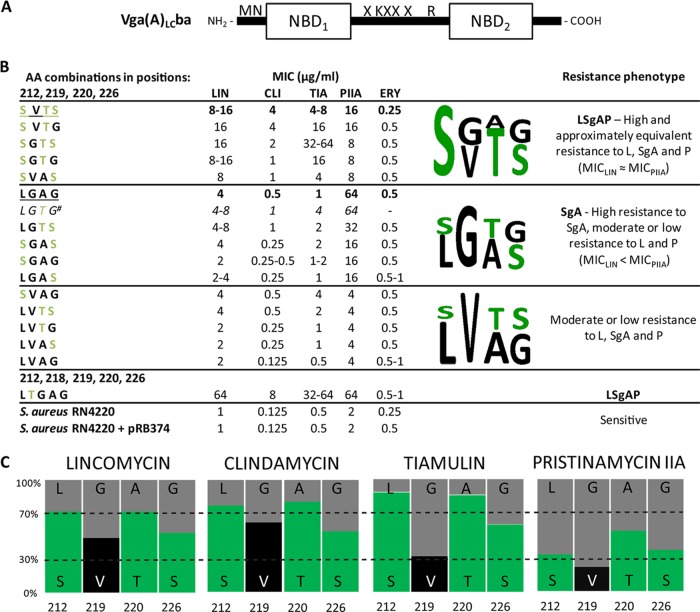
FIG 2.
Efficiency of the Vga(A)* variants prepared and analyzed in this study. (A) Scheme of the Vga(A)LCba protein used in preparing the mutant forms of Vga(A). Mutated amino acids (X) at positions 212, 219, 220, and 226 are indicated. (B) MICs, resistance phenotypes, and consensus sequence logos. The amino acid (AA) combinations at positions 212, 219, 220, and 226 are shown. The sequence patterns of the original Vga(A) and Vga(A)LC are underlined. The superscript number indicates that the predicted MIC values for the missing variant LGTG are shown. Polar amino acids are shown in green in the sequence logo. (C) Impact of the individual positions to the level of resistance to a particular antibiotic. Results are expressed as the relative ratio of the mean MIC for variants with a particular residue.