Figure 4.
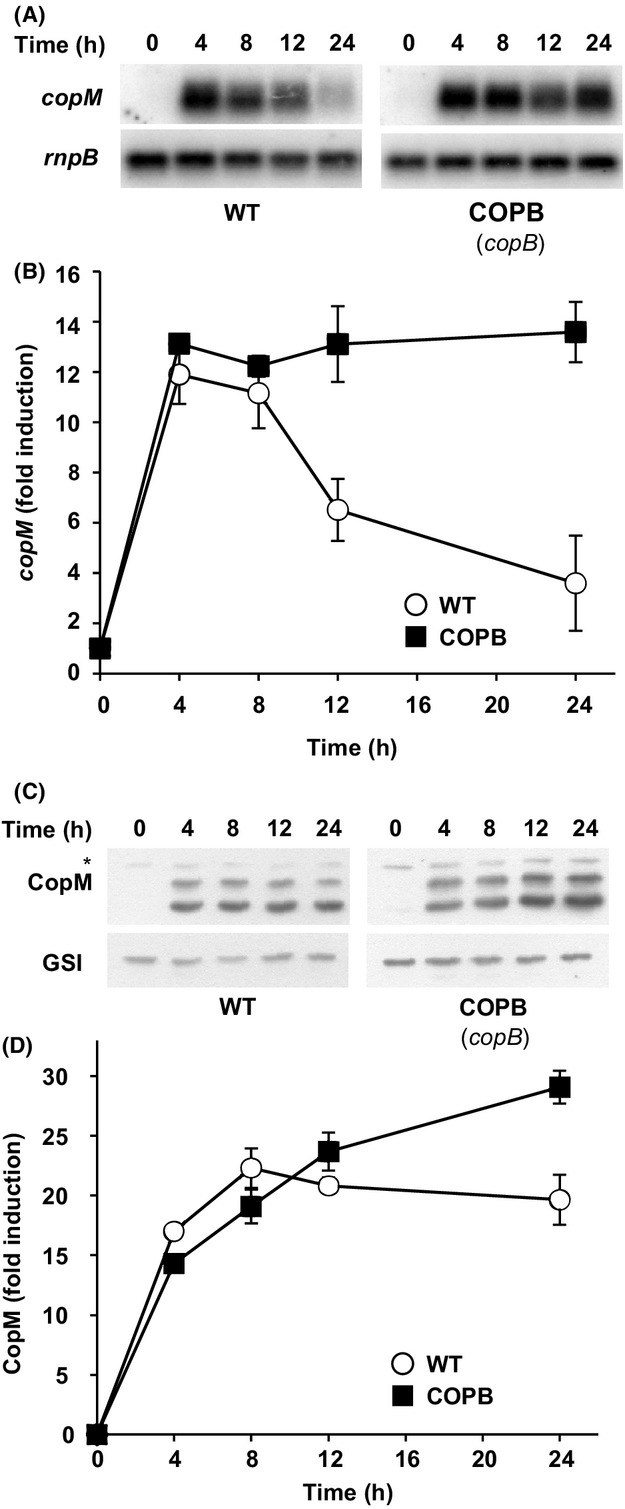
Absence of CopBAC efflux system leads to a higher accumulation of copM transcript and CopM protein. (A) Northern blot analysis of copM expression in response to copper addition in WT and COPB strains. Total RNA was isolated from WT and COPB cells grown in BG11C-Cu medium to mid-log growth phase and exposed for 24 h to copper 1 μmol/L. Samples were taken at the indicated times. The filter was hybridized with copM probe and subsequently stripped and rehybridized with an rnpB probe as a control. (B) Quantification of the relative mRNA levels of copM in response to copper addition in the WT (white circles) and the COPB (black squares) strains. Radioactive signals of three independent experiments for each strain were quantified and averaged. RNA levels were normalized with the rnpB signal. Plots of relative mRNA levels versus time were drawn; error bars represent SE. (C) Western blot analysis of CopM levels after copper addition in WT and COPB strains. Cells were grown in BG11C-Cu medium to mid-log growth phase and exposed for 24 h to copper 1 μmol/L. Five micrograms of total protein from soluble extracts was separated by 15% SDS-PAGE and analyzed by western blot to detect CopM and GSI. (D) Quantification of CopM levels in response to copper addition in WT (white circles) and COPB (black squares) strains. Western blot signal of three independent experiments were quantified using ImageJ program. CopM levels were normalized with the GSI signal. Error bars represent SE.
